Abstract
A cross-sectional study of nasopharyngeal colonization with Streptococcus pneumoniae was performed among 573 children attending 29 day-care centers (DCCs) in Norway prior to the start of mass vaccination with the heptavalent pneumococcal conjugate vaccine (PCV-7). A sensitive sampling method was employed, including transport in an enrichment broth and serotyping of pneumococci directly from the broth, in addition to traditional single-colony isolation from blood agar plates. The prevalence of carriage was high, peaking at 88.7% in 2-year-olds. More than one serotype was isolated from 12.7% of the carriers. Of 509 isolates obtained, 227 (44.6%) belonged to the PCV-7 serotypes. Penicillin nonsusceptibility was rare (1.8% of the isolates). Nonsusceptibility to erythromycin (5.9%), clindamycin (2.0%), and tetracycline (5.5%) was associated with PCV-7 serotypes (P < 0.001). Multilocus sequence typing was performed on the whole strain collection, revealing 102 sequence types (STs), of which 31 (30.4%) were novel. Eleven isolates (2.2%) belonged to the England14-9 clone, and 19 isolates (3.7%) belonged to, or were single-locus variants of, the Portugal19F-21 clone. The pneumococcal populations within the DCCs were composed of a majority of isolates with STs shared between the DCCs and a minority of isolates with STs unique for each DCC. The highest numbers of different STs, including novel STs, were found within the most frequent serotypes. Our study indicates that carriage of S. pneumoniae is highly prevalent among children in Norwegian DCCs, with a genetically diverse pneumococcal population consisting of unique microepidemic DCC populations.
Streptococcus pneumoniae (the pneumococcus) is a commensal bacterium in the human nasopharynx and a pathogen causing a wide spectrum of diseases, ranging from mild respiratory tract infections to severe invasive disease. Colonization precedes pneumococcal disease, and colonized individuals serve as a reservoir for horizontal spread of the bacterium in the community (3, 11, 18). The prevalence of asymptomatic carriage of S. pneumoniae increases during the first 2 years of life and then levels off; peak prevalences ranging from 43% to 70% have been reported in healthy children between 1 and 3 years of age in Western Europe (1, 5, 25, 26, 46).
The pneumococcus is encapsulated in a coat of polysaccharide, which gives rise to at least 90 antigenically different serotypes (20). The distribution and prevalence of serotypes differ significantly among isolates from asymptomatic carriers versus those from individuals with invasive pneumococcal disease (IPD); while some serotypes are found frequently in both conditions, others are rarely isolated from either carriage or invasive disease (19). These differences are believed to reflect in part the variable invasive potential of the serotypes (7, 39).
The emergence of antimicrobial resistance among pneumococci is a serious concern. By use of molecular typing methods, such as multilocus sequence typing (MLST), it is evident that antimicrobial-resistant pneumococci worldwide are dominated by a small number of clones (32). Currently, 26 internationally spread antibiotic-resistant clones have been described by the Pneumococcal Molecular Epidemiology Network (PMEN). These are named according to the country where they were first identified, the first serotype described, and a sequential PMEN number. Accordingly, the Portugal19F-21 clone was first identified in Portugal as a serotype 19F pneumococcus and is clone 21 in the PMEN system.
In Norway, reduced susceptibility to penicillin is rare, with 98.1% of all isolates from blood cultures being susceptible in 2006. However, resistance to erythromycin in systemic pneumococcal isolates has increased from 2.7% in 2002 to 12.4% in 2006 (33). This rapid increase in erythromycin resistance was found to be caused mainly by the spread of the England14-9 clone, carrying the mef(A) gene (29, 44).
A heptavalent conjugated pneumococcal vaccine (PCV-7) was licensed in the United States in 2000. This vaccine offers good protection against IPD caused by the included serotypes, i.e., serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F (2), and a reduction of nasopharyngeal carriage of these serotypes following vaccination has been demonstrated (10, 31). Thus, mass vaccination of children has resulted in a herd effect, as demonstrated by a reduced incidence of IPD among older adults in the United States (28). In addition, as the majority of the resistant PMEN clones have serotypes represented in PCV-7, vaccination is potentially an effective way to control the spread of resistant clones (45). However, the reduction of vaccine serotypes in asymptomatic carriage and IPD has, to some extent, been countered by replacement with nonvaccine serotypes (23, 42), requiring an improved surveillance program when PCV-7 is introduced in a population.
PCV-7 was introduced in the Norwegian childhood vaccination program in July 2006. Immunizations are administered on a three-dose schedule, with doses given at 3, 5, and 12 months of age. Because the vaccine was originally licensed for a four-dose regimen, the expected effects of the vaccination program in Norway, with respect to both carriage and IPD, might differ from those found in clinical trials and experiences from, e.g., the United States. As a consequence, surveillance of pneumococci in both carriage and disease in Norway is particularly important.
Knowledge on asymptomatic carriage of pneumococci among children attending day-care centers (DCCs) in Norway is limited. In a study performed in 2003 that included 94 children, a peak prevalence of carriage of 70% was found among children 2 years of age. Low levels of antimicrobial nonsusceptibility were revealed, with none of the isolates belonging to the PMEN clones (43). To be able to evaluate the impact of the introduction of PCV-7 in the Norwegian vaccination program, we performed a cross-sectional study to describe nasopharyngeal carriage among children attending DCCs before mass vaccination with PCV-7. The isolated pneumococci were characterized by serotyping, antimicrobial susceptibility testing, and MLST. We also evaluated factors associated with an increased risk of asymptomatic carriage.
MATERIALS AND METHODS
Study population.
In suburban areas around Oslo, the capital of Norway, children and parents/guardians in 29 DCCs were invited to participate in the present study. DCCs are generally available to the population in the area, and the majority of children attend DCCs from 1 year of age; 80.4% of 1- to 5-year-olds in Norway and 85 to 90% of 1- to 5-year-olds in the study area attend DCCs (Statistics Norway, 2006 [http://www.ssb.no]). All the parents/guardians were informed about the study by leaflets, letters, and information meetings, and written consent was obtained from all who agreed to participate. A questionnaire with data on household size, ages of family members, parents’ smoking habits, breast feeding, and use of antibiotics by the child during the last 3 months was filled out according to the information reported by the parents.
The study was approved by the Regional Committee for Medical Research Ethics, Southern Norway.
Sample collection.
Transnasal nasopharyngeal samples were collected at the DCCs between 13 September and 7 November 2006. Sampling was performed by three medical doctors; 95% of samples were taken by one of them. With the participant's head slightly tipped backwards and the tip of the nose gently lifted, a flexible wire shaft with a rayon bud (Medical Wire & Equipment, Wiltshire, United Kingdom) was inserted through a nostril, parallel to the floor of the nasal cavity, until meeting slight resistance at the posterior pharyngeal wall, about one-half to two-thirds of the distance from the nostril to the ear lobe. The swab was rotated, and preferably kept in place for 5 seconds, before removal. It was then inserted into a tube containing 3 ml of serum broth from beef infusion, which was enriched with 5% horse serum and 3.3% defibrinated horse blood (Statens Serum Institut, Copenhagen, Denmark). The swabs were transported to the laboratory and further processed within 3 to 4 h.
Bacterial identification and serotyping.
All swabs were plated onto a nonselective chocolate agar and Columbia horse blood agar containing 5.0 μg/ml gentamicin and reinserted into their respective enrichment broth used for transport. Agar plates and enrichment broths were incubated overnight at 35°C in air with 5% CO2. The presence of pneumococci was detected by direct serotyping of all enrichment cultures, as described by Kaltoft (26), using a commercial kit for latex agglutination (Pneumotest-Latex kit; Statens Serum Institut, Copenhagen, Denmark). The agglutination kit contains latex particles coated with rabbit antibodies reacting with specific pneumococcal capsular polysaccharide, and identification of pneumococcal serogroups/serotypes follows a checkerboard system after agglutination in 14 pool suspensions. Growth on the selective blood agar was examined for alpha-hemolytic colonies with typical pneumococcal morphology to confirm the findings from the broths. If more serogroups/serotypes were identified in the enrichment culture, up to 16 colonies were passaged in the attempt to identify the different strains. Pneumococci were further identified by testing for optochin sensitivity (BBL Taxo Discs; BD, NJ). The serotype was determined by the capsular reaction test (Quellung reaction) using specific antisera (Statens Serum Institut, Copenhagen, Denmark). In cases of positive agglutination of the enrichment culture but no growth on the selective blood agar, a new droplet of the enrichment broth was plated on blood agar and incubated overnight for isolation of S. pneumoniae colonies. Isolates were stored at −80°C.
Antimicrobial susceptibility testing.
For all identified isolates, antimicrobial susceptibility testing was performed by determination of the MIC using Etest (AB Biodisk, Solna, Sweden). Using the breakpoints from the Clinical and Laboratory Standards Institute (9), susceptibilities to penicillin G, cefotaxime, ceftriaxone, erythromycin, clindamycin, tetracycline, trimethoprim-sulfamethoxazole, and chloramphenicol were determined.
Genotyping.
Genomic DNA was isolated by boiling a 1-μl loopful of bacteria in 100 μl Tris-EDTA buffer for 10 min. After centrifugation at 13,000 rpm for 5 min, the supernatants were stored at −20°C. MLST was performed as described elsewhere (12). The sequence types (STs) were obtained by using the MLST database (http://www.mlst.net). Novel alleles and STs were submitted to the curator of the database and assigned designations. Clonal relationships in the strain collection were visualized using eBURST (http://eburst.mlst.net) (13). Groups of isolates sharing six of seven alleles were assigned to clonal groups (CGs).
Statistical analyses.
Analyses by the chi-square test, Fisher's exact test, and Student's t test were performed in SPSS 14.0 for Windows and GraphPad Prism 5.01. P values of <0.05 were considered significant. The genetic diversity in the total population and within each DCC was calculated using Simpson's index of diversity (D), a measure of the probability that two random and independent samples from a population will belong to the same group (41). Simpson's index of diversity is defined as 1 − λ, where λ = Σ[ni(ni − 1)]/N(N − 1), ni is the number of isolates with the ith ST in the population, and N is the number of isolates in the population.
RESULTS
Study population.
Of the 1,539 children attending the 29 DCCs, 611 (39.7%) volunteered to participate. Thirty-eight children had already received one or more doses of PCV-7. Although samples from all 611 children were obtained and analyzed, only the results for the 573 unvaccinated children were included in the analyses. The characteristics of these 573 children are shown in Table 1. The median age of the participating children was 46 months (range, 10 to 69 months); 260 (45.4%) were girls. Forty-nine children (8.6%) had received antibiotics within the last 3 months, as reported by the parents.
TABLE 1.
Characteristics of the study participants
| Characteristic | No. (%) |
|---|---|
| Age (mo) | |
| <24 | 59 (10.3) |
| 24-35 | 115 (20.1) |
| 36-47 | 132 (23.0) |
| 48-59 | 161 (28.1) |
| >60 | 106 (18.5) |
| Sex | |
| Male | 313 (54.6) |
| Female | 260 (45.4) |
| No. of persons in household | |
| 2 | 11 (11.9) |
| 3 | 105 (18.3) |
| 4 | 341 (59.5) |
| 5 | 98 (17.1) |
| >5 | 18 (3.1) |
| No. of siblings <6 yr old | |
| 0 | 286 (49.9) |
| 1 | 273 (47:6) |
| >1 | 14 (2.4) |
| Passive smoking | |
| No | 465 (81.2) |
| Yes | 108 (18.8) |
| Breast fed >6 mo | |
| No | 110 (19.2) |
| Yes | 463 (80.8) |
| RTI during past 3 mo | |
| No | 520 (90.8) |
| Yes | 53 (9.2) |
| Antibiotic use during past 3 mo | |
| No | 524 (91.4) |
| Yes | 49 (8.6) |
Prevalence of carriage and multiple colonization.
A total of 509 pneumococcal isolates were obtained from 449 (78.4%) of the 573 children. More than one strain were found in 57 children (9.9%), of which 54 (94.7%) were colonized by two strains and 3 (5.3%) were colonized by three strains. Thirty isolates (5.9%) were identified after positive broth agglutination and subsequent replating of the serum broth. Direct agglutination further indicated single-strain carriage in three children and the presence of additional strains in 19 carriers from whom one isolate had been recovered; these strains could not be grown, however, and were consequently not included in further analyses. Two pneumococcal isolates not identified by agglutination (serotypes 6B and 19F) were found during susceptibility testing due to growth within the inhibition zone; the least susceptible colonies were found to be of a different serotype than the more susceptible colonies.
Serotype distribution.
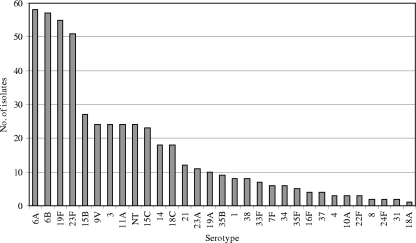
Among the 509 isolated pneumococci, 30 different serotypes were identified (Fig. 1), and 24 isolates (4.7%) were nontypeable (NT). Four serotypes, i.e., 6A, 6B, 19F, and 23F, dominated the serotype distribution by constituting 221 (43.4%) of the 509 isolates, each accounting for more than 10% of the total number of isolates. The PCV-7 serotypes, i.e., serotypes 4, 6B, 7F, 14, 18C, 19F, and 23F, constituted 227 (44.6%) of the isolates.
FIG. 1.
Distribution of serotypes among 509 S. pneumoniae isolates recovered from children attending DCCs in Norway.
Risk factor analysis.
The children colonized with pneumococci, either of any serotype or of the PCV-7 serotypes, were significantly younger than noncolonized children (P < 0.001, Student's t test). A peak prevalence of asymptomatic carriage of 88.7% was found among children aged 2 to 3 years (24 to 35 months) (Table 2). No significant associations with colonization were found for household size, number of siblings, exposure to passive smoking at home, recent upper respiratory tract infection (RTI), or recent use of antibiotics (Table 2). The duration of breast feeding was not significantly associated with colonization (P = 0.33, Student's t test).
TABLE 2.
Risk factors for colonization with S. pneumoniae in 573 children attending DCCs in Norway
| Variable | Children colonized with S. pneumoniae
|
Children colonized with PCV-7 serotype
|
||
|---|---|---|---|---|
| No. (%) | P value (chi-square test) | No. (%) | P value (chi-square test) | |
| Age (mo) | ||||
| <24 | 51 (86.4) | 0.004 | 29 (49.2) | <0.001 |
| 24-35 | 102 (88.7) | 58 (50.4) | ||
| 36-47 | 102 (77.3) | 46 (34.8) | ||
| 48-59 | 120 (74.5) | 55 (34.2) | ||
| >60 | 74 (69.8) | 27 (25.5) | ||
| Sex | ||||
| Male | 241 (77.0) | NSa | 121 (38.7) | NS |
| Female | 208 (80.0) | 94 (36.2) | ||
| No. of siblings <6 yr old | ||||
| 0 | 220 (76.9) | NS | 89 (34.3) | NS |
| 1 | 217 (79.5) | 111 (40.7) | ||
| >1 | 12 (85.7) | 7 (50.0) | ||
| Passive smoking | ||||
| No | 371 (79.8) | 0.086 | 183 (39.4) | 0.089 |
| Yes | 78 (72.2) | 33 (30.6) | ||
| RTI in past 3 mo | ||||
| No | 409 (78.7) | NS | 191 (36.7) | NS |
| Yes | 40 (75.5) | 25 (47.2) | ||
| Antibiotic use in past 3 mo | ||||
| No | 415 (79.2) | NS | 196 (37.4) | NS |
| Yes | 34 (69.4) | 20 (40.8) | ||
NS, not significant.
Antimicrobial susceptibility.
Reduced susceptibility to one or more antimicrobials was found in 105 isolates (20.6%), of which 46 (43.8%) belonged to PCV-7 serotypes (Table 3). Nine isolates(1.8%) were nonsusceptible to penicillin; one of these was additionally nonsusceptible to cefotaxime and ceftriaxone. However, no isolates were fully resistant to β-lactams. Resistance to erythromycin was found in 30 (5.9%) of the isolates; coresistance to clindamycin was found in 10 isolates (2.0%). Twenty-four isolates (4.7%) were resistant to tetracycline, and 4 isolates (0.8%) were intermediately susceptible. Nonsusceptibility to trimethoprim-sulfamethoxazole was found in 67 isolates (13.2%), with 27 (5.3%) being fully resistant and 40 (7.9%) showing intermediate susceptibility. While penicillin-nonsusceptible isolates were evenly distributed among PCV-7 serotypes (n = 5) and other serotypes (n = 4), resistance to erythromycin and tetracycline was significantly associated with PCV-7 serotypes. Of the 30 erythromycin-resistant isolates, 26 isolates (86.7%) were identified among the 227 PCV-7 serotype isolates, while 4 isolates were identified among the 282 non-PCV-7 serotypes (P < 0.001). Similarly, 26 (91.7%) of the 28 tetracycline-nonsusceptible isolates were identified among the PCV-7 serotype isolates, while 2 isolates were identified among the non-PCV-7 serotypes (P < 0.001). In contrast, nonsusceptibility to trimethoprim-sulfamethoxazole was associated with non-PCV-7 serotypes; 57 (85.1%) of 67 nonsusceptible isolates belonged to the non-PCV-7 serotypes, compared to 10 isolates among the PCV-7 serotypes (P < 0.001). Trimethoprim-sulfamethoxazole-nonsusceptible isolates were dominated by serogroup 15 and NT isolates, representing 19.4% and 17.9% of nonsusceptible isolates, respectively.
TABLE 3.
Antimicrobial susceptibilities of 509 S. pneumoniae isolates from children attending DCCs in Norway
| Antimicrobiala | No. (%) of isolates
|
No. (%) of nonsusceptible isolates belonging to a PCV-7 serotype
|
|||
|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | Intermediate | Resistant | |
| PEN | 500 (98.2) | 9 (1.8) | 5 (55.6) | ||
| ERY | 479 (94.1) | 30 (5.9) | 26 (86.7) | ||
| CLI | 499 (98.0) | 10 (2.0) | 9 (90) | ||
| TET | 481 (94.5) | 4 (0.8) | 24 (4.7) | 4 (100) | 22 (91.7) |
| SXT | 442 (86.8) | 40 (7.9) | 27 (5.3) | 6 (15) | 4 (14.8) |
PEN, penicillin; ERY, erythromycin; CLI, clindamycin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole.
Genotypes.
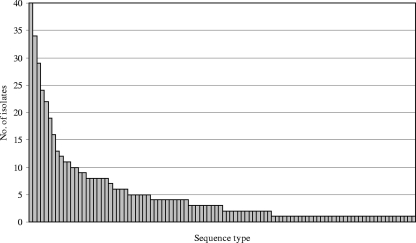
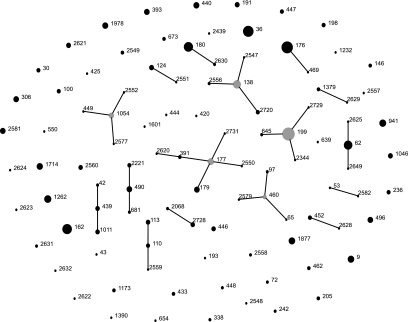
MLST was performed on all 509 isolates. Fifteen new alleles were identified and entered in the MLST database. A total of 102 STs were identified, of which 31 (30.4%) were novel STs. A small number of STs accounted for the majority of the isolates; 15 STs were recovered from at least nine or more children, and these accounted for 264 (51.9%) of the isolates (Fig. 2). These were ST199, ST176, ST36, ST162, ST180, ST62, ST138, ST1262, ST941, ST9, ST1877, ST177, ST1714, ST1978, and ST440. The most frequent ST, ST199, was identified in 40 (8.9%) of the 449 carriers. CGs consisting of groups of STs sharing alleles at six of the seven loci analyzed, i.e., single-locus variants (SLV), were assigned using eBURST. A total of 48 STs, comprising 251 (49.3%) of the isolates, clustered into 16 CGs, while 54 STs differed from other STs in more than one allele (singletons) (Fig. 3).
FIG. 2.
Distribution of STs among 509 S. pneumoniae isolates recovered from children attending DCCs in Norway. The following 15 STs, in order of decreasing frequency, were represented by nine or more isolates: ST199, ST176, ST36, ST162, ST180, ST62, ST138, ST1262, ST941, ST9, ST1877, ST177, ST1714, ST1978, and ST440.
FIG. 3.
Population snapshot of 509 S. pneumoniae isolates. An eBURST group definition of six of seven shared alleles was used. The lines connect SLVs, gray dots represent the primary founders of the CG, and numbers refer to STs. The sizes of the dots reflect the number of isolates. The PMEN clones identified are ST9 (England14-9), ST177 (Portugal19F-21), ST236 (Taiwan19F-14), ST242 (Taiwan23F-15), and ST338 (Colombia23F-26).
Genotype in relation to serotype.
For most STs represented by multiple isolates, only one serotype was identified. However, seven STs were associated with two or three serotypes. To our knowledge, five genotype-serotype associations in our material have not previously been reported: ST490 with serotype 6B, ST2068 with serotype 6A, ST1046 with serotype 34, ST1011 with serotype 23F, and ST550 with NT. These isolates were reserotyped in order to ensure that the serotype was correct. Three of these new genotype-serotype associations were found in children colonized by more than one strain: the child with the ST490 serotype 6B strain was cocolonized by an ST199 serotype 15C strain, the child with the ST1046 serotype 34 strain was cocolonized with an ST180 serotype 3 strain, and the child with the ST1011 serotype 23F strain harbored in addition both an ST2630 serotype 3 strain and an ST162 serotype 9V strain. Thus, the unusual genotype-serotype associations did not appear to result from genetic exchange between the multiple isolates evidenced in these hosts. The number of STs and the number of novel STs within a serotype increased with the number of isolates assigned to the serotype, as shown in Table 4.
TABLE 4.
Genotypes in relation to serotypes of S. pneumoniae isolates recovered from children attending DCCs in Norway
| Serotype | ST(s) (no. of isolates) | No. of:
|
||
|---|---|---|---|---|
| Isolates | STs | Novel STs | ||
| PCV-7 | ||||
| 4 | ST205 (3) | 3 | 1 | |
| 6B | ST138 (16), ST146 (3), ST176 (28), ST469 (1), ST490 (2), ST639 (1), ST2547 (1), ST2556 (1), ST2720 (4) | 57 | 9 | 3 |
| 9V | ST162 (23) | 24 | 1 | |
| 14 | ST9 (11), ST124 (6), ST2551 (1) | 18 | 3 | 1 |
| 18C | ST110 (4), ST113 (5), ST496 (8), ST2559 (1) | 18 | 4 | 1 |
| 19F | ST43 (1), ST177 (10), ST179 (8), ST236 (4), ST391 (3), ST420 (1), ST425 (1), ST462 (3), ST654 (1), ST1173 (3), ST2439 (1), ST2550 (1), ST2581 (8), ST2620 (1), ST2621 (6), ST2631 (2), ST2731 (1) | 55 | 17 | 7 |
| 23F | ST36 (29), ST242 (2), ST338 (2), ST439 (1), ST440 (9), ST1011 (5), ST2558 (2), ST2624 (1) | 51 | 8 | 2 |
| Non-PCV-7 | ||||
| 6A | ST65 (1), ST176 (6), ST460 (4), ST490 (6), ST681 (2), ST1379 (3), ST1390 (1), ST1714 (10), ST1978 (9), ST2068 (1), ST 2221 (4), ST2556(1), ST2557 (1), ST2579 (1), ST 2622 (1), ST2623 (1), ST2629 (1), ST2728 (5) | 58 | 18 | 7 |
| 18A | ST1232 (1) | 1 | 1 | |
| 19A | ST199 (6), ST645 (2), ST2548 (1), ST2632 (1) | 10 | 4 | 2 |
| 23A | ST42 (2), ST439 (4), ST2560 (5) | 11 | 3 | 1 |
| 1 | ST306 (8) | 8 | 1 | |
| 3 | ST180 (22), ST2630 (2) | 24 | 2 | 1 |
| 7F | ST191 (6) | 6 | 1 | |
| 8 | ST53 (1), ST2582 (1) | 2 | 2 | 1 |
| 10A | ST97 (2), ST2068 (1) | 3 | 2 | |
| 11A | ST62 (19), ST2549 (3), ST2625 (1), ST2649 (1) | 24 | 4 | 3 |
| 15B | ST199 (19), ST1262 (5), ST2344 (1), ST2577 (1), ST2729 (1) | 27 | 5 | 2 |
| 15C | ST199 (15), ST1262 (8) | 23 | 2 | |
| 16F | ST30 (4) | 4 | 1 | |
| 21 | ST193 (1), ST1877 (11) | 12 | 2 | |
| 22F | ST433 (3) | 3 | 1 | |
| 24F | ST72 (2) | 2 | 1 | |
| 31 | ST444 (1), ST1601 (1) | 2 | 2 | |
| 33F | ST100 (4), ST673 (3) | 7 | 2 | |
| 34 | ST1046 (6) | 6 | 1 | |
| 35B | ST198 (4), ST452 (4), ST2628 (1) | 9 | 3 | 1 |
| 35F | ST446 (5) | 5 | 1 | |
| 37 | ST447 (4) | 4 | 1 | |
| 38 | ST393 (8) | 8 | 1 | |
| NT | ST448 (2), ST449 (1), ST550 (1), ST941 (12), ST1054 (7), ST2552 (1) | 24 | 6 | 1 |
Antimicrobial nonsusceptibility and genotypes.
A total of 32 STs were identified among the nonsusceptible isolates, of which 7 were novel STs (Table 5). Five STs belonged to one of the 26 worldwide-spread resistant pneumococcal clones currently accepted by PMEN (i.e., England14-9, Taiwan19F-14, Taiwan23F-15, Portugal19F-21, and Colombia23F-26), accounting for 26 (24.8%) of the 105 nonsusceptible isolates, and three STs were SLVs of PMEN clones. However, in one of these PMEN STs, ST236, only one of four isolates were nonsusceptible to antibiotics. The majority of STs unrelated to the PMEN clones included isolates with monoresistance to trimethoprim-sulfamethoxazole. Thirteen STs (ST43, ST100, ST113, ST176, ST193, ST440, ST460, ST941, ST1046, ST1262, ST1978, ST2557, and ST2560), accounting for 51.4% of the nonsusceptible isolates, were nonsusceptible solely to trimethoprim-sulfamethoxazole.
TABLE 5.
Characteristics of 105 isolates of S. pneumoniae that are not susceptible to antibiotics
| ST | Serotype | No. of nonsusceptible isolates/total no. of isolates | Antimicrobial nonsusceptibilitya | Clone |
|---|---|---|---|---|
| ST9 | 14 | 11/11 | ERY | England14-9 |
| ST43 | 19F | 1/1 | SXT | |
| ST62 | 11A | 3/19 | ERY, SXT | |
| ST100 | 33F | 1/4 | SXT | |
| ST110 | 18C | 1/4 | PEN, ERY | |
| ST113 | 18C | 1/5 | SXT | |
| ST162 | 9V | 1/23 | ERY, SXT | Spain9V-3 SLV |
| ST176 | 6B | 1/34 | SXT | |
| ST177 | 19F | 10/10 | TET | Portugal19F-21 |
| ST179 | 19F | 8/8 | ERY, CLI, TET | Portugal19F-21 SLV |
| ST193 | 21 | 1/1 | SXT | |
| ST199 | 19A | 1/40 | PEN, SXT | |
| ST236 | 19F | 1/4 | PEN, ERY, TET, SXT | Taiwan19F-14 |
| ST242 | 23F | 2/2 | ERY, TET, SXT | Taiwan23F-15 |
| ST338 | 23F | 2/2 | PEN | Colombia23F-26 |
| ST440 | 23F | 2/9 | SXT | |
| ST460 | 6A | 1/4 | SXT | |
| ST462 | 19F | 3/3 | TET | |
| ST469 | 6B | 1/1 | ERY | |
| ST490 | 6A | 1/8 | ERY, CLI, TET | |
| ST550 | NT | 1/1 | PEN, SXT | |
| ST941 | NT | 12/12 | SXT | |
| ST1046 | 34 | 6/6 | SXT | |
| ST1262 | 15B/C | 13/13 | SXT | |
| ST1978 | 6A | 9/9 | SXT | |
| ST2550 | 19F | 1/1 | TET | Portugal19F-21 SLV |
| ST2557 | 6A | 1/1 | SXT | |
| ST2560 | 23A | 5/5 | SXT | |
| ST2577 | 15B | 1/1 | TET | |
| ST2622 | 6A | 1/1 | PEN, TET | |
| ST2624 | 23F | 1/1 | PEN, ERY, CLI, TET, SXT | |
| ST2632 | 19A | 1/1 | PEN, CTX, CRO, SXT |
PEN, penicillin; CTX, cefotaxime; CRO, ceftriaxone; ERY, erythromycin; CLI, clindamycin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole.
Resistance to erythromycin, clindamycin, and tetracycline are often associated. Of the 45 isolates that were nonsusceptible to one of these substances in this study, 29 (64.4%) belonged to two CGs: ST9, the England14-9 clone with erythromycin monoresistance, and CG177, including ST177 and ST179 (the Portugal19F-21 clone and an SLV), showing resistance to tetracycline and tetracycline plus macrolides, respectively.
The penicillin-nonsusceptible isolates were heterogeneous; six different capsule phenotypes (6A, 18C, 19A, 19F, 23F, and NT) and eight STs, including three novel STs, were identified. Two STs belonged to the PMEN clones. The most extensive drug resistance was found in a serotype 23F isolate, ST2624, showing intermediate susceptibility to penicillin and resistance to erythromycin, clindamycin, tetracycline, and trimethoprim-sulfamethoxazole. The only isolate with nonsusceptibility to penicillin and cephalosporins was a serotype 19A isolate of the novel ST2632.
Multiple colonization.
Among the isolates recovered from the 57 children colonized by more than one strain, the most prevalent types were 19F, 3, NT, 6A, 6B, 23F, 15C, and 9V, with each of them represented by more than five isolates. Three STs were identified more than five times in these carriers, i.e., ST180, ST162, and ST199, representing 11, 6, and 6 isolates, respectively. No pattern of association between the serotypes or STs of the isolates was discernible. Isolates of different serotypes carried simultaneously in these 57 children had ≤3 common alleles by MLST, indicating genetic unrelatedness. Serotype 3, however, was positively associated with multiple carriage; of the 24 serotype 3 isolates identified, 13 were recovered from the 57 carriers of multiple serotypes, while 11 were recovered from the 392 carriers of one serotype (P < 0.001).
Distribution of genotypes in DCCs.
Of the 102 STs, 51 (50%) were found in more than one DCC, accounting for 425 (83.5%) isolates, and 51 (50%) were restricted to one DCC, accounting for 84 (16.5%) isolates. Of the 31 novel STs, 26 were restricted to only one DCC; 22 of these were recovered from only one child. In 15 DCCs, the pneumococcal population was dominated by one or two STs recovered from more than four children (Table 6). Among these dominating STs, five were frequently recovered overall in the study, while the remaining nine STs, including two novel STs (ST2560 and ST2581) were rarely or never found outside the DCC where they dominated. The STs dominating a DCC represented up to 55.6% of the isolates (DCC VII).
TABLE 6.
Genotypic diversity of S. pneumoniae in 29 DCCs in Norway
| DCC | No. of:
|
ST(s) with >4 isolates (no. of isolates) | Simpson's index of diversitya | |||
|---|---|---|---|---|---|---|
| Children sampled | Children colonized | Isolates | STs | |||
| I | 31 | 24 | 24 | 9 | 199 (10) | 0.81 |
| II | 30 | 24 | 25 | 11 | 36 (8) | 0.88 |
| III | 19 | 15 | 15 | 8 | 176 (6) | 0.84 |
| IV | 30 | 23 | 25 | 10 | 62 (5), 199 (6) | 0.89 |
| V | 30 | 18 | 20 | 10 | 1978 (8) | 0.82 |
| VI | 20 | 13 | 16 | 8 | 439 (5) | 0.88 |
| VII | 16 | 14 | 18 | 8 | 941 (10) | 0.70 |
| VIII | 22 | 19 | 22 | 14 | 0.96 | |
| IX | 11 | 9 | 9 | 8 | 0.97 | |
| X | 10 | 9 | 11 | 7 | 179 (5) | 0.82 |
| XI | 24 | 15 | 17 | 11 | 0.91 | |
| XII | 28 | 22 | 29 | 13 | 162 (9) | 0.89 |
| XIII | 17 | 15 | 17 | 10 | 162 (7) | 0.84 |
| XIV | 18 | 18 | 22 | 8 | 393 (8) | 0.83 |
| XV | 9 | 7 | 7 | 7 | 1.0 | |
| XVI | 7 | 7 | 7 | 3 | 0.64 | |
| XVII | 17 | 14 | 18 | 12 | 0.92 | |
| XVIII | 22 | 22 | 29 | 18 | 2560 (5) | 0.96 |
| XIX | 22 | 18 | 21 | 8 | 1046 (6) | 0.87 |
| XX | 31 | 22 | 22 | 14 | 0.95 | |
| XXI | 6 | 6 | 6 | 5 | 0.86 | |
| XXII | 30 | 23 | 23 | 11 | 0.92 | |
| XXIII | 23 | 15 | 16 | 11 | 0.95 | |
| XXIV | 20 | 19 | 21 | 11 | 0.93 | |
| XXV | 22 | 15 | 19 | 10 | 2581 (6) | 0.87 |
| XXVI | 16 | 13 | 16 | 9 | 0.88 | |
| XXVII | 12 | 8 | 8 | 5 | 0.92 | |
| XXVIII | 14 | 12 | 14 | 6 | 9 (5) | 0.81 |
| XXIX | 16 | 10 | 11 | 7 | 0.87 | |
Simpson's index of diversity (D) = 1 − Σ[ni(ni − 1)]/N(N − 1), where ni is the number of isolates with the ith ST in the population and N is the number of isolates in the population.
The genotypic diversity of the total population of carriage isolates was 0.973, as measured by Simpson's index of diversity, D. The genotypic diversity within DCCs ranged from 0.64 to 1.0, the latter indicating a population from a DCC in which each single isolate had a different ST (Table 6).
DISCUSSION
In this study of pneumococcal carriage in children attending DCCs in Norway, a high overall carriage rate of 78.4% was found, with a peak prevalence of 88.7% in children 2 to 3 years old. These frequencies are higher than previously described in Norway, as well as in similar studies performed in other Scandinavian countries or elsewhere in Western Europe (5, 43, 46). This is probably due to the highly sensitive sampling method used in this study, i.e., giving the bacteria optimal conditions in an enrichment broth directly after sampling, allowing a short interval before plating, and performing serotyping directly from the incubated enrichment broth to identify multiple serotypes. The use of serum broth as a sensitive method for recovery of pneumococci has been described by Kaltoft (26). Detection of capsular antigen after enrichment has been demonstrated as a sensitive way to identify pneumococci in carriage studies, yielding more than culture on agar plates alone and with no further gain from adding gentamicin in the enrichment broth (27). In the present study, the presence of pneumococci was indicated by agglutination in 33 samples that were initially culture negative on blood agar. In 30 of these samples, growth was obtained after replating of the enrichment broth, increasing the yield by 7.2%, from 419 to 449 positive samples. The lack of recovery of strains from three antigen-positive samples might be due to a small amount of bacteria in the sample, autolysis of the pneumococci or overgrowth by other species. Hence, transport in an enrichment broth, followed by incubation and serotyping directly from the broth, is a sensitive method for isolation of pneumococci. However, the method is suitable only when plating can be performed within a short time interval after sample collection.
Longitudinal studies have demonstrated that children are successively colonized with multiple strains of pneumococci (18). However, identification of simultaneous carriage of multiple serotypes is laborious, and the yield varies according to the method used (24). Serotyping of multiple colonies is time-consuming, and if the second serotype constitutes 4 to 27% of the population, 11 to 59 randomly picked CFU would have to be subcultured and serotyped for identification of the two serotypes (24). An immunoblot method designed to detect multiple serotypes in carriage studies was used in a study in Navajo and White Mountain Apache reservations. Multiple serotypes were detected in 8.1% of positive samples (6, 34). In a study of pneumococcal carriage in the Gambia, serotyping was performed by latex agglutination, and multiple serotypes were identified in 11.5% of positive samples (30). We performed a serotype screening by latex agglutination on incubated enrichment broths, followed by subcultivation of up to 16 colonies from blood agar to isolate multiple strains. By this method, more than one serotype was recovered in 57 (12.7%) of 449 positive specimens. Additional serotypes were indicated in 19 positive specimens by broth agglutination, but these organisms were not identified on agar plates, possibly due to small fractions of the pneumococcal populations being constituted by these strains. It is possible that additional pneumococcal strains in a sample might be lost as a result of negative agglutination. In fact, two isolates were identified as additional strains as a result of differing inhibition zones on susceptibility testing. No agglutination-negative samples showed growth on blood agar, indicating that agglutination is highly sensitive. Direct agglutination from an enrichment broth thus has the advantage of being an easy and inexpensive method for identification of more than one pneumococcal strain per sample. By this method, random colony selection can be limited to samples where carriage of multiple serotypes is indicated by agglutination.
Crowding of children in DCCs gives ample opportunities for transmission of bacteria, and DCC attendance is considered a strong risk factor for carriage of pneumococci (37). This high-risk setting might in part explain the high prevalence of carriage found in this study. Nasopharyngeal sampling was performed in the autumn, shortly after the summer holidays and at the beginning of a new season in the DCCs with new children attending. This might be a vulnerable time point for the acquisition of pneumococci because of a disturbance of the DCC epidemiological unit and good opportunities for exchange of bacteria between the children not yet immunologically adapted to this new environment. The prevalence of carriage was significantly associated with low age, as was colonization with PCV-7 serotypes, an association that has been documented by others (22, 37). Although RTIs and young siblings have been identified as risk factors and use of antibiotics and breast feeding have been negatively associated with carriage (14, 37), no statistically significant associations with these risk factors were found in this study. This is not surprising, as the population studied is believed to be very homogenous and considering the high prevalence of carriage, making the identification of potential risk factors for carriage problematic.
The distribution of serotypes is consistent with results of other studies of nasopharyngeal carriage, with serotypes 6B, 6A, 19F, and 23F being the most prevalent (4, 16, 25, 46). The proportion of PCV-7 serotypes among isolated pneumococci was 45%. This proportion is consistent with results from The Netherlands (42%) and from a previous study from Norway (42%), although a higher PCV-7 coverage (64%) has been reported in the United Kingdom (4, 25, 43). The potential coverage of an 11-valent vaccine (including serotypes 1, 3, 5, and 7F) would be only slightly higher (47.3%).
In our study, 8 isolates (1.6%) belonged to serotype 1. This serotype is associated with epidemic outbreaks of pneumococcal disease, though it is rarely recovered from asymptomatic carriers (8). The prevalence of serotype 1 pneumococci increased in Scandinavian countries during the 1990s and accounted for 11.1% of cases of systemic pneumococcal disease in children aged 0 to 5 years in Norway in 1995 to 2001, making it the third most prevalent serotype causing invasive disease in children (36). This increase was primarily due to the emergence of one antibiotic-susceptible clone, ST306 (21), a clone that seems to be genetically stable and that was found to be the most prevalent serotype 1 clone in Europe in the past decade (8). All serotype 1 pneumococci isolated in the present study belonged to this ST. Serotype 1 pneumococci are rare in the United States, and this serotype is not included in PCV-7. Thus, the selective pressure caused by mass vaccination in Norway might have implications for the future occurrence of this serotype.
To our knowledge, this is the largest study of carriage in which MLST has been performed on all isolates. The genotyping of 509 isolates showed that the pneumococcal population was highly heterogeneous; 102 STs were identified, and the most frequent contributed to fewer than 8% of the isolates. Of the 15 clones most frequently recovered in this study, all have previously been described in other countries, including 2 of the 26 worldwide-spread resistant pneumococcal clones currently accepted by PMEN, England14-9 (ST9) and Portugal19F-21 (ST177). Nearly one-third of the STs were novel, the majority of which were recovered from only one child. The proportion of novel STs, and the variation of STs, was highest among the most frequently recovered serotypes, i.e., 6A, 6B, 19F, and 23F. These serotypes are carried for longer time periods than other serotypes (18), and they might have a greater opportunity for horizontal transfer of genetic material, giving rise to new STs. However, this could also be influenced by a sampling bias, with the highest clonal diversity being found among the most frequently recovered serotypes. The novel STs might represent a Norwegian pneumococcal population. However, as the majority of these novel clones were recovered from only a few children each, they might be clones with a limited success in transmission.
The antimicrobial susceptibility among pneumococci recovered in Norway is favorable; the isolates are generally sensitive to the drugs tested. Penicillin nonsusceptibility was found in a small fraction of isolates, i.e., nine isolates (1.8%) belonging to eight clones, of which four displayed PCV-7 serotypes. The isolates that were nonsusceptible to macrolides and tetracycline were more homogenous and were assigned to a few CGs displaying PCV-7 serotypes. Thus, a reduction, or limited spread, of these clones is expected after the introduction of the conjugate vaccine in the Norwegian vaccination program.
The rapid increase in macrolide resistance observed in Norway among IPD isolates from 2001 and onwards was attributable mainly to the England14-9 clone, displaying low-level resistance to erythromycin (44). However, from 2004, the fraction of pneumococci with coresistance to erythromycin and clindamycin, indicating a macrolide-lincosamine-streptogramin B-type resistance, increased, and in 2006 this phenotype constituted 15% of erythromycin-nonsusceptible invasive isolates in Norway (33). Among the pneumococci recovered in this study, 10 (33%) of the erythromycin-nonsusceptible isolates were coresistant to clindamycin. Of these, eight isolates (80%) belonged to the Portugal19F-21 CG. In addition, this phenotype was found in one of the eight serotype 6A, ST490 isolates recovered in this study; a clone unrelated to the PMEN clones and not previously associated with this resistance pattern. The macrolide-lincosamine-streptogramin B phenotype was also found in a novel ST (ST2624), represented by the most extensively nonsusceptible isolate in this material, which was intermediately susceptible to penicillin and resistant to erythromycin, clindamycin, tetracycline, and trimethoprim-sulfamethoxazole. CG177, consisting of Portugal19F-21 and its SLVs, constituted 19 (18.1%) of the nonsusceptible isolates in this study. These isolates display serotype 19F, a serotype included in PCV-7. However, this serotype has been associated with vaccine failures (2), and in a community-randomized trial, carriage of this serotype has actually been found to be higher among children who had received four doses of PCV-7 than in controls (34). The effects of a three-dose vaccination regimen on the carriage and spread of these clones need to be followed closely.
The clone most frequently isolated from the children was ST199, displaying serotypes 15B, 15C, and 19A. Increasing prevalences of IPD caused by the nonvaccine serotypes 19A, 15B, and 15C in the United States have been described in the years following licensure of PCV-7 (17, 35). This replacement of PCV-7 serotypes in IPD has occurred 3 to 4 years after implementation of widespread vaccination and has been found to be mainly due to an expansion of CG199, i.e., ST199 and newly described SLVs (35). Although the invasiveness of this clone is believed to be moderate (7), its high prevalence among asymptomatic carriers in Norway, already before the introduction of PCV-7, raises concern that a similar replacement in IPD might occur.
Multiple pneumococcal strains simultaneously carried by the individual children were genetically unrelated to each other, as shown by MLST. However, serotype 3 pneumococci were isolated at a significantly higher frequency from carriers of multiple serotypes than from the total population, with ST180 being the most frequently identified ST from multiple carriage. Serotype 3 has been found to be positively associated with acute otitis media and acute conjuncitivitis (40), but with a low potential for causing invasive disease (7). It is possible that the capsule of serotype 3 is favorable for colonization of mucosal surfaces, but the reason for its overrepresentation in cocolonization with other pneumococci is unclear. The mucoid appearance of serotype 3 colonies might make them more discernible in a mixed population and might bias this observation. However, it is possible that this particular serotype has a strong genetic integrity and ability to survive in the competition with other pneumococci for colonization of the nasopharyngeal niche.
Pneumococci recovered within a single DCC were genotypically less diverse than the total pneumococcal population, and one or two dominating clones were identified in most DCCs. The composition of pneumococcal populations within each DCC consisted of the dominating clones, either predominantly found in that DCC or recovered from several DCCs, and of clones, often novel STs, unique for the DCC. In this way the clonal distribution of pneumococci was unique for each DCC, a pattern described as an autonomous epidemiological unit by Sa-Leao et al. (38). The observed distribution of clones within DCCs, as contrasted to the total population, is consistent with the suggested model of the emergence of a neutral bacterial population structure from overlapping microepidemics within clustered host populations (15). However, longitudinal studies of DCC populations are needed to evaluate the stability of the DCC epidemiological unit, as a cross-sectional study will catch only a glimpse of a probably highly dynamic interplay between children and the pneumococci.
The high prevalence of pneumococcal carriage among children in DCCs described in this study underlines the importance of this population as a reservoir for spread of the bacterium to the whole community. In addition, the high level of DCC attendance among Norwegian children might augment the importance of these epidemiological settings in the spread of pneumococci. Consequently, the reduced colonization, and the resulting herd immunity, is an advantage of conjugate vaccination. To evaluate the effect of a three-dose PCV-7 immunization regimen on carriage and spread of susceptible and nonsusceptible clones of S. pneumoniae, a follow-up study is planned for 2008.
Acknowledgments
This study was supported in part by grant 06_14 from the Norwegian surveillance system for antimicrobial resistance (NORM).
We thank Fredrik Næss for contributing to the collection of nasopharyngeal samples. We thank Anne R. Alme, Torill Alvestad, Anne M. Klem, May-Liss Knudsen, Janne O. Rønning, and Gunnhild Rødal for excellent technical assistance.
Footnotes
Published ahead of print on 4 June 2008.
REFERENCES
- 1.Aniansson, G., B. Alm, B. Andersson, P. Larsson, O. Nylen, H. Peterson, P. Rigner, M. Svanborg, and C. Svanborg. 1992. Nasopharyngeal colonization during the first year of life. J. Infect. Dis. 165S38-S42. [DOI] [PubMed] [Google Scholar]
- 2.Black, S., H. Shinefield, B. Fireman, E. Lewis, P. Ray, J. R. Hansen, L. Elvin, K. M. Ensor, J. Hackell, G. Siber, F. Malinoski, D. Madore, I. Chang, R. Kohberger, W. Watson, R. Austrian, K. Edwards, et al. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Pediatr. Infect. Dis. J. 19189-195. [DOI] [PubMed] [Google Scholar]
- 3.Bogaert, D., R. de Groot, and P. W. Hermans. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4144-154. [DOI] [PubMed] [Google Scholar]
- 4.Bogaert, D., M. Sluijter, N. L. Toom, T. J. Mitchell, W. H. Goessens, S. C. Clarke, R. de Groot, and P. W. Hermans. 2006. Dynamics of pneumococcal colonization in healthy Dutch children. Microbiology 152377-385. [DOI] [PubMed] [Google Scholar]
- 5.Bogaert, D., A. van Belkum, M. Sluijter, A. Luijendijk, R. de Groot, H. C. Rumke, H. A. Verbrugh, and P. W. Hermans. 2004. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 3631871-1872. [DOI] [PubMed] [Google Scholar]
- 6.Bronsdon, M. A., K. L. O'Brien, R. R. Facklam, C. G. Whitney, B. Schwartz, and G. M. Carlone. 2004. Immunoblot method to detect Streptococcus pneumoniae and identify multiple serotypes from nasopharyngeal secretions. J. Clin. Microbiol. 421596-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 1871424-1432. [DOI] [PubMed] [Google Scholar]
- 8.Brueggemann, A. B., and B. G. Spratt. 2004. Geographic distribution and clonal diversity of Streptococcus pneumoniae serotype 1 isolates. J. Clin. Microbiol. 1901211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. M100-S17. Clinical Laboratory Standards Institute, Wayne, Pa.
- 10.Dagan, R., R. Melamed, M. Muallem, L. Piglansky, D. Greenberg, O. Abramson, P. M. Mendelman, N. Bohidar, and P. Yagupsky. 1996. Reduction of nasopharyngeal carriage of pneumococci during the second year of life by a heptavalent conjugate pneumococcal vaccine. J. Infect. Dis. 1741271-1278. [DOI] [PubMed] [Google Scholar]
- 11.De Lencastre, H., and A. Tomasz. 2002. From ecological reservoir to disease: the nasopharynx, day-care centres and drug-resistant clones of Streptococcus pneumoniae. J. Antimicrob. Chemother. 5075-81. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 1443049-3060. [DOI] [PubMed] [Google Scholar]
- 13.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkelstein, J. A., S. S. Huang, J. Daniel, S. L. Rifas-Shiman, K. Kleinman, D. Goldmann, S. I. Pelton, A. DeMaria, and R. Platt. 2003. Antibiotic-resistant Streptococcus pneumoniae in the heptavalent pneumococcal conjugate vaccine era: predictors of carriage in a multicommunity sample. Pediatrics 112862-869. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, C., W. P. Hanage, and B. G. Spratt. 2005. Neutral microepidemic evolution of bacterial pathogens. Proc. Natl. Acad. Sci. USA 1021968-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldblatt, D., M. Hussain, N. Andrews, L. Ashton, C. Virta, A. Melegaro, R. Pebody, R. George, A. Soininen, J. Edmunds, N. Gay, H. Kayhty, and E. Miller. 2005. Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: a longitudinal household study. J. Infect. Dis. 192387-393. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez, B. E., K. G. Hulten, L. Lamberth, S. L. Kaplan, E. O. Mason, Jr., and the U.S. Pediatric Multicenter Pneumococcal Surveillance Group. 2006. Streptococcus pneumoniae serogroups 15 and 33: an increasing cause of pneumococcal infections in children in the United States after the introduction of the pneumococcal 7-valent conjugate vaccine. Pediatr. Infect. Dis. J. 25301-305. [DOI] [PubMed] [Google Scholar]
- 18.Gray, B. M., G. M. Converse III, and H. C. Dillon, Jr. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142923-933. [DOI] [PubMed] [Google Scholar]
- 19.Hausdorff, W. P., D. R. Feikin, and K. P. Klugman. 2005. Epidemiological differences among pneumococcal serotypes. Lancet Infect. Dis. 583-93. [DOI] [PubMed] [Google Scholar]
- 20.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 332759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henriques Normark, B., M. Kalin, A. Ortqvist, T. Akerlund, B. O. Liljequist, J. Hedlund, S. B. Svenson, J. Zhou, B. G. Spratt, S. Normark, and G. Kallenius. 2001. Dynamics of penicillin-susceptible clones in invasive pneumococcal disease. J. Infect. Dis. 184861-869. [DOI] [PubMed] [Google Scholar]
- 22.Hill, P. C., A. Akisanya, K. Sankareh, Y. B. Cheung, M. Saaka, G. Lahai, B. M. Greenwood, and R. A. Adegbola. 2006. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian villagers. Clin. Infect. Dis. 43673-679. [DOI] [PubMed] [Google Scholar]
- 23.Huang, S. S., R. Platt, S. L. Rifas-Shiman, S. I. Pelton, D. Goldmann, and J. A. Finkelstein. 2005. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics 116e408-e413. [DOI] [PubMed] [Google Scholar]
- 24.Huebner, R. E., R. Dagan, N. Porath, A. D. Wasas, and K. P. Klugman. 2000. Lack of utility of serotyping multiple colonies for detection of simultaneous nasopharyngeal carriage of different pneumococcal serotypes. Pediatr. Infect. Dis. J. 191017-1020. [DOI] [PubMed] [Google Scholar]
- 25.Hussain, M., A. Melegaro, R. G. Pebody, R. George, W. J. Edmunds, R. Talukdar, S. A. Martin, A. Efstratiou, and E. Miller. 2005. A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol. Infect. 133891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaltoft, M. S. 2003. Pneumococcal carriage among children attending day care centres and invasive pneumococcal infections in childhood. Ph.D. thesis. University of Aarhus, Aarhus, Denmark.
- 27.Lankinen, K. S., P. Salo, S. Rapola, E. Salo, A. K. Takala, and M. Leinonen. 1997. Pneumococcal capsular antigen detection after enrichment culture: an alternative to culture methods in epidemiologic research. Am. J. Trop. Med. Hyg. 56211-215. [DOI] [PubMed] [Google Scholar]
- 28.Lexau, C. A., R. Lynfield, R. Danila, T. Pilishvili, R. Facklam, M. M. Farley, L. H. Harrison, W. Schaffner, A. Reingold, N. M. Bennett, J. Hadler, P. R. Cieslak, and C. G. Whitney. 2005. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. JAMA 2942043-2051. [DOI] [PubMed] [Google Scholar]
- 29.Littauer, P., M. Sangvik, D. A. Caugant, E. A. Høiby, G. S. Simonsen, and A. Sundsfjord. 2005. Molecular epidemiology of macrolide-resistant isolates of Streptococcus pneumoniae collected from blood and respiratory specimens in Norway. J. Clin. Microbiol. 432125-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd-Evans, N., T. J. O'Dempsey, I. Baldeh, O. Secka, E. Demba, J. E. Todd, T. F. Mcardle, W. S. Banya, and B. M. Greenwood. 1996. Nasopharyngeal carriage of pneumococci in Gambian children and in their families. Pediatr. Infect. Dis. J. 15866-871. [DOI] [PubMed] [Google Scholar]
- 31.Mbelle, N., R. E. Huebner, A. D. Wasas, A. Kimura, I. Chang, and K. P. Klugman. 1999. Immunogenicity and impact on nasopharyngeal carriage of a nonavalent pneumococcal conjugate vaccine. J. Infect. Dis. 1801171-1176. [DOI] [PubMed] [Google Scholar]
- 32.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 392565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.NORM/NORM-VET. 2007. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. National Veterinary Institute, Tromsø/Oslo, Norway.
- 34.O'Brien, K. L., E. V. Millar, E. R. Zell, M. Bronsdon, R. Weatherholtz, R. Reid, J. Becenti, S. Kvamme, C. G. Whitney, and M. Santosham. 2007. Effect of pneumococcal conjugate vaccine on nasopharyngeal colonization among immunized and unimmunized children in a community-randomized trial. J. Infect. Dis. 1961211-1220. [DOI] [PubMed] [Google Scholar]
- 35.Pai, R., M. R. Moore, T. Pilishvili, R. E. Gertz, C. G. Whitney, B. Beall, and Active Bacterial Core Surveillance Team. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 1921988-1995. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen, M. K., E. A. Høiby, L. O. Frøholm, V. Hasseltvedt, G. Lermark, and D. A. Caugant. 2004. Systemic pneumococcal disease in Norway 1995-2001: capsular serotypes and antimicrobial resistance. Epidemiol. Infect. 132167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regev-Yochay, G., M. Raz, R. Dagan, N. Porat, B. Shainberg, E. Pinco, N. Keller, and E. Rubinstein. 2004. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin. Infect. Dis. 38632-639. [DOI] [PubMed] [Google Scholar]
- 38.Sa-Leao, R., A. Tomasz, I. S. Sanches, S. Nunes, C. R. Alves, A. B. Avo, J. Saldanha, K. G. Kristinsson, and H. De Lencastre. 2000. Genetic diversity and clonal patterns among antibiotic-susceptible and -resistant Streptococcus pneumoniae colonizing children: day care centers as autonomous epidemiological units. J. Clin. Microbiol. 384137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandgren, A., K. Sjostrom, B. Olsson-Liljequist, B. Christensson, A. Samuelsson, G. Kronvall, and N. B. Henriques. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189785-796. [DOI] [PubMed] [Google Scholar]
- 40.Shouval, D. S., D. Greenberg, N. Givon-Lavi, N. Porat, and R. Dagan. 2006. Site-specific disease potential of individual Streptococcus pneumoniae serotypes in pediatric invasive disease, acute otitis media and acute conjunctivitis. Pediatr. Infect. Dis. J. 25602-607. [DOI] [PubMed] [Google Scholar]
- 41.Simpson, E. H. 1949. Measurement of diversity. Nature 163688. [Google Scholar]
- 42.Singleton, R. J., T. W. Hennessy, L. R. Bulkow, L. L. Hammitt, T. Zulz, D. A. Hurlburt, J. C. Butler, K. Rudolph, and A. Parkinson. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 2971784-1792. [DOI] [PubMed] [Google Scholar]
- 43.Sogstad, M. K., I. S. Aaberge, J. O. Sordal, E. A. Høiby, L. O. Frøholm, A. R. Alme, and D. A. Caugant. 2006. Carriage of Streptococcus pneumoniae in healthy Norwegian children attending day-care centres. Eur. J. Clin. Microbiol. Infect. Dis. 25510-514. [DOI] [PubMed] [Google Scholar]
- 44.Sogstad, M. K., P. Littauer, I. S. Aaberge, D. A. Caugant, and A. Høiby. 2007. Rapid spread in Norway of an erythromycin-resistant pneumococcal clone, despite low usage of macrolides. Microb. Drug Resist. 1329-36. [DOI] [PubMed] [Google Scholar]
- 45.Stephens, D. S., S. M. Zughaier, C. G. Whitney, W. S. Baughman, L. Barker, K. Gay, D. Jackson, W. A. Orenstein, K. Arnold, A. Schuchat, M. M. Farley, and Georgia Emerging Infections Program. 2005. Incidence of macrolide resistance in Streptococcus pneumoniae after introduction of the pneumococcal conjugate vaccine: population-based assessment. Lancet 365855-863. [DOI] [PubMed] [Google Scholar]
- 46.Syrjanen, R. K., T. M. Kilpi, T. H. Kaijalainen, E. E. Herva, and A. K. Takala. 2001. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J. Infect. Dis. 184451-459. [DOI] [PubMed] [Google Scholar]