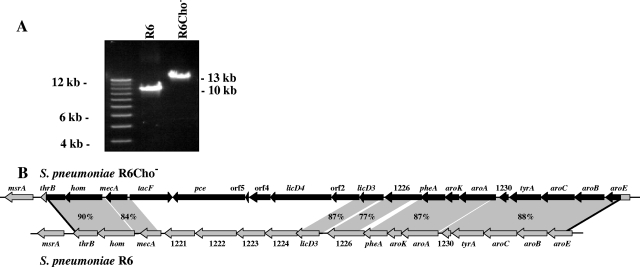
FIG. 3.
(A) PCR analysis of the licD3 locus in S. pneumoniae R6 and the choline-independent derivative R6Cho−. Long-range PCR products using primers located in hom and pheA (see panel B) with genomic DNA of R6 and R6Cho− are shown on a 1% agarose gel. (B) Schematic representation of the licD3 locus in S. pneumoniae R6 and the choline-independent derivative R6Cho−. The genomic organization of the licD3 region of S. pneumoniae R6 (bottom) is shown along with the same region of S. pneumoniae R6Cho− after integration of S. oralis ATCC 35037 DNA (top). Endogenous R6 genes are shown by gray arrows and are indicated by gene names or “spr” numbers (11, 12, 31). Acquired S. oralis genes are shown by black arrows. The sites of recombination are indicated by black lines. Replacement of S. pneumoniae R6 DNA by S. oralis ATCC 35037 DNA occurred between thrB (position 1.214.973 in the R6 genome) and aroE (position 1.231.893). The degree of identity (percent) between S. pneumoniae and S. oralis genes is indicated. Some of the gene products, whose genes showed no significant similarity to R6, could be identified by their protein similarity. Designations orf2, orf4, and orf5 were given according to their positions in the presumed licD3 operon of S. oralis. Identities of R6 genes spr1221 to spr1224 to S. oralis genes could not be assessed because the whole-genome sequence of S. oralis ATCC 35037 is not available. S. oralis homologues of R6 genes spr1226 and spr1230 were given the same number to highlight similarity. They are not likely to occupy the same position in the S. oralis genome. The S. oralis nucleotide sequence is available from GenBank (accession number EU675999).