Abstract
Tetrocarcin A (TCA), produced by Micromonospora chalcea NRRL 11289, is a spirotetronate antibiotic with potent antitumor activity and versatile modes of action. In this study, the biosynthetic gene cluster of TCA was cloned and localized to a 108-kb contiguous DNA region. In silico sequence analysis revealed 36 putative genes that constitute this cluster (including 11 for unusual sugar biosynthesis, 13 for aglycone formation, and 4 for glycosylations) and allowed us to propose the biosynthetic pathway of TCA. The formation of d-tetronitrose, l-amicetose, and l-digitoxose may begin with d-glucose-1-phosphate, share early enzymatic steps, and branch into different pathways by competitive actions of specific enzymes. Tetronolide biosynthesis involves the incorporation of a 3-C unit with a polyketide intermediate to form the characteristic spirotetronate moiety and trans-decalin system. Further substitution of tetronolide with five deoxysugars (one being a deoxynitrosugar) was likely due to the activities of four glycosyltransferases. In vitro characterization of the first enzymatic step by utilization of 1,3-biphosphoglycerate as the substrate and in vivo cross-complementation of the bifunctional fused gene tcaD3 (with the functions of chlD3 and chlD4) to ΔchlD3 and ΔchlD4 in chlorothricin biosynthesis supported the highly conserved tetronate biosynthetic strategy in the spirotetronate family. Deletion of a large DNA fragment encoding polyketide synthases resulted in a non-TCA-producing strain, providing a clear background for the identification of novel analogs. These findings provide insights into spirotetronate biosynthesis and demonstrate that combinatorial-biosynthesis methods can be applied to the TCA biosynthetic machinery to generate structural diversity.
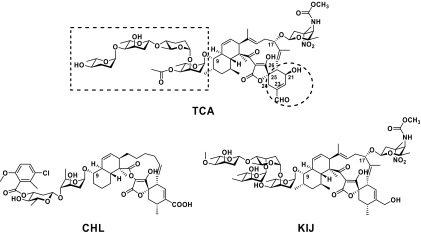
Tetrocarcin A (TCA), produced by Micromonospora chalcea NRRL 11289, belongs to a growing family of spirotetronate antibiotics with both antibacterial and antitumor activities (37, 42, 43). As shown in Fig. 1, TCA possesses an unusual polycyclic aglycone (tetronolide) that features a trans-decalin system and a tetronate moiety spiro-linked with a cyclohexene ring. To furnish the molecule, tetronolide has two sugar side chains appended, a tetrasaccharide unit that comprises alternate l-digitoxoses and l-amicetoses and 2,3,4,6-tetradeoxy-4-(methylcarbamoyl)-3-C-methyl-3-nitro-d-xylo-hexopyranose, a rare deoxynitrosugar also known as d-tetronitrose or d-kijanose in kijanimicin (KIJ) (21, 39). Intriguingly, recent studies have demonstrated that TCA can induce apoptosis of various tumor cells in a cell-type-dependent manner, e.g., by (i) antagonizing the mitochondrial functions of proteins of the Bcl-2 family (natural apoptosis inhibitors) in HeLa cells (8, 26), (ii) activating the caspase-dependent cell death pathway via endoplasmic reticulum stress in lymphomas (1, 41), and (iii) inhibiting the phosphorylation of factors involved in phosphatidylinositol 3-kinase/Akt signaling in human breast cancer cells (25). Synthetic TCA analogs that showed the separable antibacterial and antitumor activities also supported the versatile modes of action of TCA in vivo (12). Upon bioassay using experimental mouse models, TCA exhibited remarkable antitumor activities without significant myelosuppression and nephrotoxicity associated (24), suggesting that it could serve as a promising chemotherapeutic agent for further evaluation in preclinical trials.
FIG. 1.
Structures of TCA, CHL, and KIJ. TCA has an aglycone similar to those of CHL and KIJ but shows differences mainly in the functionalities of the cyclohexene ring (in the dashed circle) and a glycosyl substitution at the C-9 position (in the dashed rectangle).
The rich and complex functionalities of the spirotetronate architecture pose an attractive but tremendous challenge to chemical synthesis. Since the mid 1980s, extensive attempts to synthesize aglycones have been made, resulting in the total synthesis of (+)-tetronolide of TCA and (−)-chlorothricolide of chlorothricin (CHL) (the first member to be discovered and structurally elucidated in this family [Fig. 1]) (4, 30, 31, 36). However, the synthesis of the entire molecule of either TCA or CHL has not yet been achieved. On the other hand, investigation of the structure-biological activity relationship for a better understanding of the mechanisms and pharmaceutical development of TCA mainly relies on its structural derivatization. Synthetic modifications of TCA have been limited to exchange of the sugar side chains (12), and alterations of the aglycone have not been explored, again probably due to the structural complexity.
Combinatorial biosynthesis, which aims at expanding the structural diversity of complex natural products by genetic engineering of their biosynthetic pathways, offers a promising alterative way to solve this problem. For the success of this approach, the biosynthetic mechanisms of spirotetronate antibiotics need to be elucidated. Additionally, TCA is one of the members containing the richest and most diverse unusual sugars (i.e., one tetronitrose, two amicetoses, and two digitoxoses) in the spirotetronate family. Most members have similar glycosylation positions but show distinct patterns in chain length and sugar units. Alterations in the tetrasaccharide side chain remarkably affected the biological activities of TCA (12), suggesting that glycodiversification at the C-9 position could serve as a targeting factor for further drug development. Characterization of genes governing the biosynthesis of tetronitrose, amicetose, and digitoxose will not only provide new insights into unusual sugar biosynthesis, but also contribute to the general field of combinatorial biosynthesis by enriching the “toolbox” for glycodiversification (3, 7, 20, 32).
Previous isotope-labeled feeding experiments showed that the aglycone of TCA is primarily of polyketide origin (39), except for C-24 to C-26. A similar labeling pattern was found in the biosynthetic study of CHL (10, 22), and the three remaining carbons at corresponding positions were confirmed to be derived from the glycerol (17), suggesting that a common strategy involves the incorporation of a glycerol-derived 3-C unit with a polyketide chain to form the aglycones in spirotetronate biosynthesis. In 2006, we reported the cloning and characterization of the chl gene cluster as the first genetic model to propose the biosynthetic pathway in this family (11). Combined with the very recently proposed biosynthetic pathways of KIJ and the polyether tetronomycin (TMN) containing a tetronate moiety and biochemical characterization of the first enzymatic step toward the tetronate moiety in TMN (5, 35, 44), the deduced biosynthetic mechanisms for spirotetronate aglycones are becoming clear but are far from being determined.
Based on the structural similarity between TCA and CHL and the polyketide origin revealed by isotope-labeling experiments, we report access to the TCA biosynthetic machinery in M. chalcea NRRL 11289 by heterologous hybridization, using a set of CHL type I polyketide synthase (PKS) gene fragments as mixed probes. The cloning and sequencing of the entire tca gene cluster allowed the assignment of functions to the deduced gene products, providing insights into the biosynthetic pathway of TCA. Sequence comparison of this gene cluster to those for CHL, KIJ, and TMN led to identification of the set of unique genes tcaD1 to tcaD4 for tetronate moiety biosynthesis. In this subpathway, in vitro characterization of the first enzymatic step by utilization of 1,3-biphosphoglycerate (1,3-BPG) as the substrate for 3-C unit incorporation and in vivo cross-complementation of the bifunction-fused gene tcaD3, respectively, to ΔchlD3 and ΔchlD4 in CHL biosynthesis strongly supported a highly conserved strategy to form this characteristic spirotetronate in tetronate-containing natural-product biosynthesis. The TCA biosynthetic machinery is amenable to genetic manipulation. Deletion of the large DNA fragment harboring PKS genes resulted in a non-TCA-producing strain, providing a clear background as a useful negative control for identification of unknown related compounds from the wild-type strain and novel analogs from the recombinant strains generated by genetic engineering of the TCA biosynthetic pathway.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used in this study are summarized in Table 1. Biochemicals, including coenzyme A (CoA), d-3-phosphoglyceric acid disodium salt (d-3-PG) and 3-phosphoglyceric phosphokinase (PGK), were purchased from the Sigma-Aldrich Co. (St. Louis, MO); chemicals and media were from the Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China), or Oxoid Ltd. (Basingstoke, United Kingdom); and restriction enzymes were from the Dalian Takara Biotechnology Co., Ltd. (Dalian, China).
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain/plasmid | Characteristics | Source/reference |
|---|---|---|
| E. coli | ||
| DH5α | Host for general cloning | Invitrogen |
| VCS257 | Host for construction of the genomic library in pOJ446 | Strategene |
| EPI300-T1R | Host for construction of the genomic library in Cloning-Ready CopyControl | Epicentre |
| S17-1 | Donor strain for conjugation between E. coli and M. chalcea | 21 |
| BL21(DE3) | Host for protein expression | Novagen |
| M. chalcea | ||
| NRRL 11289 | Wild-type strain; TCA producing | NRRL |
| TL3003 | pks deletion mutant that lacks partial tcaA1 and A5 and entire tcaA2 and A4; non-TCA producing | This study |
| S. antibioticus | ||
| DSM 40725 | Wild-type strain; CHL producing | DSMZ |
| TL1006 | ΔchlD3 mutant; non-CHL producing | 11 |
| TL1007 | ΔchlD4 mutant; non-CHL producing | 11 |
| TL3001 | TL1006 derivative containing pTL3015; CHL producing | This study |
| TL3002 | TL1007 derivative containing pTL3015; CHL producing | This study |
| Plasmids | ||
| pOJ446 | E. coli-Streptomyces shuttle cosmid for construction of the genomic library | NRRL 14791 |
| pCC1FOS | Fosmid for construction of the genomic library | Epicentre |
| pGEM-5zf | Subcloning vector in E. coli | Promega |
| pSP72 | Subcloning vector in E. coli | Promega |
| pANT841 | Subcloning vector in E. coli | GenBank (accession no. AF438749) |
| pBluescript SK(+) | Subcloning vector in E. coli | Strategene |
| pET28a | Vector for protein expression in E. coli | Novagen |
| pQ6 | Vector for protein expression in E. coli | Unpublished |
| pOJ260 | E. coli-Streptomyces shuttle vector; nonreplicating in Streptomyces | NRRL 14785 |
| pTGV2 | E. coli-Streptomyces shuttle vector for heterologous expression | Unpublished |
| pTGE33 | pET28a derivative for expression of the phosphopantetheinyl transferase (Sfp) | Unpublished |
| pTL3001 | pOJ446-based M. chalcea genomic library cosmid | This study |
| pTL3002 | pOJ446-based M. chalcea genomic library cosmid | This study |
| pTL3003 | pOJ446-based M. chalcea genomic library cosmid | This study |
| pTL3004 | pCC1FOS-based M. chalcea genomic library fosmid | This study |
| pTL3005 | pCC1FOS-based M. chalcea genomic library fosmid | This study |
| pTL3006 | pCC1FOS-based M. chalcea genomic library fosmid | This study |
| pTL3007 | pCC1FOS-based M. chalcea genomic library fosmid | This study |
| pTL3008 | 1.9-kb PCR product containing chlD1 in pBluescript SK(+) | This study |
| pTL3009 | pET28a derivative for expression of ChlD1 in E. coli | This study |
| pTL3010 | 0.2-kb PCR product containing chlD2 in pBluescript SK(+) | This study |
| pTL3011 | pET28a derivative for expression of ChlD2 in E. coli | This study |
| pTL3012 | 0.2-kb PCR product containing tcaD2 in pSP72 | This study |
| pTL3013 | pQ6 derivative for expression of TcaD2 in E. coli | This study |
| pTL3014 | 2.1-kb PCR product containing tcaD3 in pGEM-5zf | This study |
| pTL3015 | 2.1-kb EcoRI/HindIII fragment that contains tcaD3 under the control of PermE* in pTGV2 | This study |
| pTL3016 | 6.0-kb BamHI/NotI fragment from pTL3001 in pANT841 | This study |
| pTL3017 | 4.5-kb BamHI/NotI fragment from pTL3002 in pANT841 | This study |
| pTL3018 | pOJ260 derivative; construct for deletion of a large DNA region (30.146 kb) harboring pks genes (including partial tcaA1 and tcaA5 and intact tcaA2 and tcaA4) in M. chalcea | This study |
DNA isolation, manipulation, and sequencing.
DNA isolation and manipulation in Escherichia coli and M. chalcea were carried out according to standard methods (14, 33). PCR amplifications were carried out on an authorized thermal cycler (AG 22331; Eppendorf, Hamburg, Germany) using either Taq DNA polymerase or PrimStar High-Fidelity DNA polymerase (Dalian Takara Biotechnology Co., Ltd., Dalian, China). Primer synthesis and general DNA sequencing were carried out at the Shanghai Invitrogen Biotechnology Co., Ltd. (Shanghai, China). The sequencing of cosmids and fosmids was performed on the 3730xl DNA analyzer (Applied Biosystems Co.) at the Chinese National Human Genome Center (Shanghai, China), using a shotgun cloning strategy.
Genomic-library construction and screening.
The first genomic library of M. chalcea NRRL 11289 was constructed in pOJ446 according to a method described previously (19). This library was screened by colony hybridization with mixed heterologous probes, the labeled PCR products P1, P2, and P4 that were previously used for identification of the chl gene cluster (11). The resultant positive clones were further confirmed by Southern hybridization.
To extend the cloned DNA region on the chromosome, the second genomic library of M. chalcea was constructed in CopyControl pCC1FOS according to the manufacturer's instructions (catalog no. CCFOS110; Epicentre Technologies Co.). This library was screened by colony hybridization with probes P5 (a 2.7-kb NcoI/XbaI fragment) from cosmid pTL3001 and P6 (a 4.0-kb NcoI/SpeI fragment) from cosmid pTL3002, respectively. The resultant positive clones were further confirmed by Southern hybridization.
In silico sequence analysis.
The open reading frames (ORFs) were deduced from the sequence with the FramePlot 4.0beta program (http://nocardia.nih.go.jp/fp4). The corresponding deduced proteins were compared with other known proteins in the database by available BLAST methods (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Amino acid sequence alignments were performed by the CLUSTALW method from Biology WorkBench 3.2 software (http://workbench.sdsc.edu). Identification of functional domains of PKSs was carried out by using a program provided at the website http://www.nii.res.in/searchall.html.
Generation of holo-ACP from apo-ACP.
For phosphopantetheinylation of acyl carrier proteins (ACPs), each reaction was carried out in 100 μl of 50 mM Tris-HCl buffer (pH 7.5) containing 10 mM MgCl2, 1 mM Tris-(2-carboxylethyl)-phosphine (TCEP), 0.3 mM CoA, 5 μM phosphopantetheinyl transferase Sfp from Bacillus subtilis (27), and 150 μM apo-ChlD2 or apo-TcaD2 at 30°C for 1 h. Control reactions were performed in the absence of Sfp. To quench the reaction, 20 μl of 10% formic acid was added.
Generation of glyceroyl-S-ACP from holo-ACP.
For the generation of 1,3-BPG in situ, each reaction was carried out in 200 μl of 50 mM Tris-HCl buffer (pH 7.5) containing 10 mM MgCl2, 1 mM TCEP, 5 mM ATP, 0.25 mM d-3-PG, 50 U PGK, and 2.5 μM ChlD1 at 30°C for 5 to 10 min. For transfer of the glyceroyl group, 50 μl of the above-mentiioned reaction mixture that contained holo-ChlD2 or holo-TcaD2 was added, and the 250 μl of combined mixture was further incubated at 30°C for 5 min, 10 min, 15 min, 20 min, 30 min, or 60 min. Control reactions were performed in the absence of d-3-PG, ChlD1, PGK, or ChlD2/TcaD2. To quench the reaction, 50 μl of 10% formic acid was added.
Analysis of protein samples.
High-performance liquid chromatography (HPLC) analysis was carried out on a GraceVydac protein and peptide C18 column (catalog no. 218TP54; serial no. E030813-1B-2; W. R. Grace & Co., Connecticut). The column was equilibrated with 80% solvent A (H2O, 0.1% formic acid) and 20% solvent B (CH3CN, 0.1% formic acid) and developed with the following program: 0 to 3 min, constant 80% A/20% B; 3 to 5 min, a linear gradient to 60% A/40% B; 5 to 23 min, a linear gradient to 42% A/58% B; 23 to 25 min, a linear gradient to 10% A/90% B; and 25 to 30 min, a linear gradient to 80% A/20% B. This was carried out at a flow rate of 1 ml/min with UV detection at 220 nm, using a Jasco 2010 HPLC system (Jasco Co., Ltd., Japan). Each product fraction was collected and concentrated and then subjected to matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) (AutoFlex MALDI-TOF-MS; Bruker Daltonics Inc.) analysis at the Research Center of Proteome Analysis, Chinese Academy of Sciences (Shanghai, China). Protein samples were agitated with the matrix α-cyano-4-hydroxycinnamic acid and then irradiated by the N2 lasing light emitter at 337 nm to generate the detectable cations.
Production, isolation, and analysis of CHL in Streptomyces antibioticus.
The production, isolation, and HPLC analysis of CHL were described previously (11). To confirm the identity of CHL, HPLC-MS (for HPLC, Agilent 1100 [Agilent Technologies Co.]; for MS, Thermo Fisher LTQ Fleet [Thermo Fisher Scientific Inc.]) analysis was carried out, exhibiting an [M-H]− ion at an m/z of 953.5, consistent with the molecular formula C50H63ClO16.
Production, isolation, and analysis of TCA in M. chalcea.
For fermentation, each 50 μl of mycelium suspension was inoculated into 3 ml of tryptic soy broth and incubated at 28°C and 220 rpm for 36 h. One milliliter of this culture was transferred into a 250-ml flask containing 50 ml of seed medium (1% glucose, 2.4% soluble starch, 0.3% beef extract, 0.5% tryptone, 0.5% yeast extract, 2.0% CaCO3, pH 7.2) (38) and incubated at 28°C and 220 rpm for 36 to 48 h. A total of 10 ml of seed culture was then inoculated into 100 ml of fermentation medium (6% soluble starch, 1% dry yeast, 1% β-cyclodextrin, 0.2% CaCO3, pH 6.8) in a 500-ml flask and incubated at 28°C and 220 rpm for 64 to 72 h.
For isolation, each 100 ml of the culture broth was centrifuged for 5 min at 15,554 × g for removal of the precipitate. The supernatant was adjusted to a pH of 2.0 and extracted three times with 120 ml of ethyl acetate. The combined extract was then concentrated in a vacuum and resolved in 1 ml of methanol.
HPLC analysis was carried out on a Waters Cosmosil C18 column (4.6 by 250 mm; product no. K57965; code no. 38145-21). The column was equilibrated with 40% solvent A (H2O, 0.1% formic acid) and solvent B (CH3CN, 0.1% formic acid) and developed with the following program: 0 to 5 min, constant 60% A/40% B; 5 to 16 min, a linear gradient from 60% A/40% B to 20% A/80% B; 16 to 25 min, constant 20% A/80% B; 25 to 30 min, a linear gradient from 20% A/80% B to 60% A/40% B. This was carried out at a flow rate of 1 ml/min with UV detection at 216 nm, using an Agilent 1100 HPLC system. The identity of TCA was confirmed by HPLC-MS analysis performed under the same conditions. TCA showed an [M+NH4]+ ion at an m/z of 1,330.0, consistent with the molecular formula C67H96N2O24.
Supporting information.
For supporting data, including MALDI-TOF MS analysis of ACPs, HPLC-MS analysis of quasimolecular and fragmentary ions, protein expression and purification, heterologous complementation, and PKS gene deletion in this study, see the supplemental material.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in GenBank under accession number EU443633.
RESULTS AND DISCUSSION
Cloning and sequencing of the TCA biosynthetic gene cluster from M. chalcea NRRL 11289.
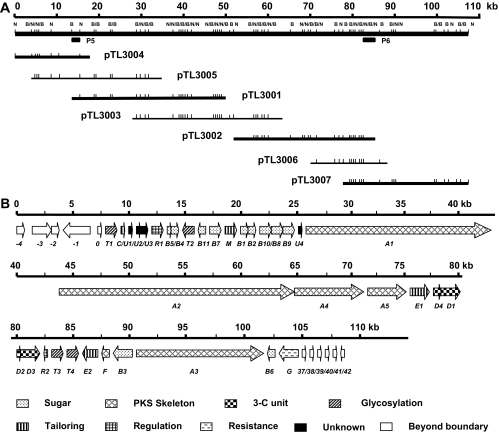
The similarities in both structures and polyketide origins between CHL and TCA indicate that conserved PKS systems may primarily govern the biosynthesis of their aglycones. Accordingly, several PKS gene fragments from the CHL biosynthetic gene cluster were chosen as heterologous probes for initial screening of the pOJ446-based cosmid library of M. chalcea NRRL 11286. The mixed probes contained the PCR products P1, P2, and P4 that were used for identification of the chl gene cluster and were confirmed by sequencing to be fragments from the loci of chlA2, chlA1, and chlA5, respectively, which cover an approximately 50-kb DNA region encoding the CHL type I PKSs (11). Upon screening of ∼6,000 clones, 30 overlapping cosmids that spanned a 70- to 72-kb DNA region were identified. To ensure full coverage of the entire tca gene cluster, additional chromosome walking was carried out using the 2.7-kb NcoI/XbaI fragment P5 from cosmid pTL3001 toward the left side and the 4.0-kb NcoI/SpeI fragment P6 from pTL3002 toward the right side (Fig. 2A). Although several attempts failed to extend the cloned region on the pOJ446-based cosmid library, screening of a newly constructed fosmid library of M. chalcea led to the identification of 18 fosmids and eventually resulted in an approximately 110-kb contiguous DNA region on the chromosome, as exemplified by the inserts of cosmids pTL3001, pTL3002, and pTL3003 and fosmids pTL3004, pTL3005, pTL3006, and pTL3007 (Fig. 2A).
FIG. 2.
Cloned DNA region on the chromosome of M. chalcea, restriction map, and genetic organization of the tca gene cluster. (A) DNA locus represented by three cosmids and four fosmids, and restriction map. The solid black bars indicate regions whose sequences have been determined. The solid quadrates indicate the probes' loci (P5, a 2.7-kb NcoI/XbaI fragment from pTL3001; P6, a 4.0-kb NcoI/SpeI fragment from pTL3002). B, BamHI; N, NcoI. (B) A 108-kb sequenced DNA region from the cosmids pTL3001 and pTL3002 and fosmids pTL3004 and pTL3007 and genetic organization of the tca gene cluster. The proposed functions of individual ORFs are labeled and are summarized in Table 2.
The DNA region represented by cosmids pTL3001 and pTL3002 and fosmids pTL3004 and pTL3007 was selected for sequencing, yielding a 108,236-bp contiguous sequence with 72.9% of the overall GC content, characteristic of actinomycete DNA. Bioinformatic analysis of the sequenced region revealed 47 ORFs (Fig. 2B and Table 2), 36 of which, from tcaT1 to tcaG, were proposed to constitute the tca gene cluster according to the functional assignment of their deduced products and genetic comparison to the gene clusters for CHL and KIJ biosynthesis. The genes beyond this region lacked significant similarities to those for secondary-metabolite biosynthesis.
TABLE 2.
Deduced functions of ORFs in the TCA biosynthetic gene cluster
| Gene | Sizea | Protein, homolog,b and origin | % Identity/similarity | Proposed function |
|---|---|---|---|---|
| orf(−4)c | 134 | CBG1971 (CAE72536.1); Caenorhabditis briggsae | 36/47 | Hypothetical protein |
| orf(−3)c | 296 | CitA (ZP_00616833.1); Kineococcus radiotolerans SRS30216 | 52/63 | ATP-binding region, ATPase-like |
| orf(−2)c | 186 | CitB (NP_629572.1); Streptomyces coelicolor A3(2) | 58/67 | Two-component response regulator |
| orf(−1)c | 1,031 | DnaE (ZP_01650120.1); Salinispora arenicola CNS205 | 87/93 | DNA polymerase III, alpha subunit |
| orf(0)c | 105 | Unknown protein | ||
| tcaT1 | 370 | STIAU_3104 (ZP_01461407); Stigmatella aurantiaca DW4/3-1 | 30/42 | GT |
| TcaC | 186 | RHA1_ro09012 (YP_708214); Rhodococcus sp. RHA1 | 55/68 | Flavin reductase |
| tcaU1 | 109 | Strop_3510 (5059984); Salinispora tropica CNB-440 | 65/69 | Unknown protein |
| tcaU2 | 146 | CLOL250_02534 (EDO56846); Clostridium sp. strain L2-50 | 32/56 | Unknown protein |
| tcaU3 | 312 | Unknown protein | ||
| tcaR1 | 267 | ChlF2 (AAZ77687); S. antibioticus | 44/59 | Pathway-specific activator |
| tcaB5 | 197 | TylCVII (AAD41825); Streptomyces fradiae | 60/73 | Sugar (3,)5-epimerase |
| tcaB4 | 332 | ChlC4 (AAZ77681); S. antibioticus | 56/69 | Sugar 3-ketoreductase |
| tcaT2 | 392 | ChlC7 (AAZ77672); S. antibioticus | 37/53 | GT |
| tcaB11 | 263 | Methyltransferase (ZP_01650637); S. arenicola CNS205 | 38/53 | SAM-dependent N-methyltransferase |
| tcaB7 | 380 | SpnR (AAG23279); Sacharopolyspora spinosa SpnQ (AAG23278); S. spinosa | 55/71 26/50 | Sugar aminotransferase/3,4-dehydratase |
| TcaM | 380 | Srm6 (CM96572); Streptomyces ambofaciens | 40/56 | O-Acyltransferase |
| tcaB1 | 295 | AclY (AAK83289); Steptomyces galilaeus | 62/75 | TDP-d-glucose synthase |
| tcaB2 | 332 | AprE (AAG18457); Streptomyces tenebrarius | 74/83 | TDP-glucose 4,6-dehydratase |
| tcaB10 | 434 | RubN8 (CAI94696); Streptomyces achromogenes subsp. Rubradiris | 61/74 | Oxidoreductase |
| tcaB8 | 373 | Med-ORF20 (BAC79028); Streptomyces sp. strain AM-7161 | 80/86 | Sugar 3-aminotransferase |
| tcaB9 | 414 | PCZA361.22 (CAA11777); Amycolatopsis orientalis | 69/79 | Sugar 3-methyltransferase |
| tcaU4 | 164 | YD repeat (ZP_00573146); Frankia sp. strain EAN1pec | 41/52 | PAS/PAC sensor hybrid histidine kinase |
| tcaA1 | 6,330 | FRAAL4072 (YP_714269); Frankia alni ACN14a | 44/55 | Type I polyketide synthase |
| tcaA2 | 7,007 | FscC (AAQ82564); Streptomyces sp. strain FR-008 | 49/61 | Type I polyketide synthase |
| tcaA4 | 1,840 | HbmAII (AAY28226); Streptomyces hygroscopicus | 49/62 | Type I polyketide synthase |
| tcaA5 | 1,537 | ChlA6 (AAZ77699); S. antibioticus | 51/62 | Type I polyketide synthase |
| tcaE1 | 491 | ChlE3 (AAZ77700); S. antibioticus | 48/61 | FAD-dependent oxidoreductase |
| tcaD4 | 350 | ChlM (AAZ77702); S. antibioticus | 65/76 | Ketosynthase III-like condensation protein |
| tcaD1 | 633 | ChlD1 (AAZ77703); S. antibioticus | 60/70 | Acyltransferase and phosphotase |
| tcaD2 | 75 | ChlD2 (AAZ77704); S. antibioticus | 57/72 | ACP |
| tcaD3 | 616 | N-terminal domain to ChlD3 (AAZ77705); S. antibioticus C-terminal domain to ChlD4 (AAZ77706); S. antibioticus | 58/74 55/71 | Dihydrolipoamide acyltransferase and hydrolase |
| tcaR2 | 192 | SACE_3583 (YP_001105782); Sacharopolyspora erythraea NRRL 2338 | 40/56 | TetR family transcriptional regulator |
| tcaT3 | 398 | ChlC7 (AAZ77672); S. antibioticus | 41/55 | GT |
| tcaT4 | 379 | ChlC7 (AAZ77672); S. antibioticus | 42/57 | GT |
| tcaE2 | 505 | MitR (AAD28454); Streptomyces lavendulae | 49/65 | Flavin-dependent oxidoreductase |
| TcaF | 254 | SAV407 (AAD28454); S. avermitilis MA-4680 | 54/70 | Thioesterase |
| tcaB3 | 487 | ORF26 (AAP85341); Streptomyces griseoruber | 53/64 | Sugar 2,3-dehydratase |
| tcaA3 | 3,964 | ChlA5 (AAZ77698); S. antibioticus | 55/66 | Type I polyketide synthase |
| tcaB6 | 289 | SCO3730 (NP_627921); S. coelicolor A3(2) | 59/71 | NAD(P)H-dependent aldo/keto reductase |
| TcaG | 490 | ChlG (AAZ77686); S. antibioticus | 43/61 | Multidrug export protein |
| orf37c | 275 | STIAU_2740 (ZP_01466190.1); Stigmatella aurantiaca DW4/3-1 | 39/53 | M23 peptidase domain protein |
| orf38c | 222 | CHAP (ZP_00532045.1)Chlorobium phaeobacteroides BS1 | 32/52 | Integrin alpha chain |
| orf39c | 132 | mll5401 (NP_106072.1); Mesorhizobium loti MAFF303099 | 40/52 | Hypothetical protein |
| orf40c | 106 | ArsR (YP_001135161.1); Mycobacterium gilvum PYR-GCK | 78/84 | Regulatory protein |
| orf4c | 243 | MMT1 (YP_001135160.1); M.m gilvum PYR-GCK | 76/83 | Co/Zn/Cd cation transporter-like protein |
| orf42c | 148 | PldB (YP_286985.1); Dechloromonas aromatica RCB | 41/57 | Alpha/beta hydrolase fold |
The size of each protein is shown as the number of amino acids.
NCBI accession numbers are given in parentheses.
ORF beyond the tca gene cluster.
Genes for regulation and resistance.
Two genes, tcaR1 and tcaR2 (Fig. 2B), are presumed to encode the pathway-specific regulatory proteins within the tca gene cluster. TcaR1 contains a putative effector domain of a response regulator at the N terminus and a transcriptional activator domain at the C terminus, suggesting that it may function as a regulatory member of a two-component system to positively control TCA production. TcaR2 belongs to a TetR family of transcriptional regulators that are widely found in microorganisms. Only one resistance candidate, tcaG, which encodes a putative efflux protein, was identified in the tca gene cluster.
Genes encoding the biosynthesis of deoxy(nitro)sugar moieties.
Deoxysugars, which are frequently structural components of natural products, are important, in many cases essential, for biological activities (9). TCA contains five deoxysugar units, including two l-amicetoses, two l-digitoxoses, and one d-tetronitrose. The last two are also found in the KIJ structure, forming the sugar side chains at the C-9 and C-17 positions, respectively. At least 11 genes, tcaB1 to tcaB11, were identified and proposed to be responsible for the biosynthesis of these unusual sugars, as outlined in Fig. 3A. The mechanisms unique to l-amicetose biosynthesis and important differences between TCA and KIJ sugar genes are described below.
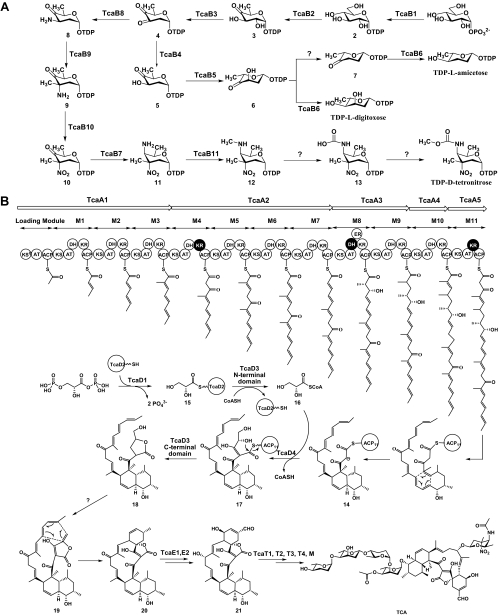
FIG. 3.
Proposed biosynthetic pathway of TCA. (A) Pathways for d-tetronitrose, l-amicetose, and l-digitoxose. (B) Mode for aglycone assembly, glycosylations, and tailoring modifications. 1, glucose-1-phosphate; 2, TDP-d-glucose; 3, TDP-6-deoxy-4-keto-d-glucose; 4, TDP-2,6-dideoxy-3,4-diketo-d-glucose; 5, TDP-2,6-dideoxy-4-keto-d-glucose; 6, TDP-2,6-dideoxy-4-keto-l-allose; 7, TDP-2,3,6-trideoxy-4-keto-l-hexose; 8, TDP-2,3,6-trideoxy-4-keto-3-amino-d-glucose; 9, TDP-2,3,6-trideoxy-4-keto-3-amino-3-C-methyl-d-hexose; 10, TDP-2,3,6-trideoxy-4-keto-3-C- methyl-3-nitro-d-hexose; 11, TDP-2,3,4,6-tetrodeoxy-4-amino-3-C-methyl-3-nitro-d-hexose; 12, TDP-2,3,4,6-tetrodeoxy-4-methylamino-3-C-methyl-3-nitro-d-hexose; 13, TDP-2,3,4,6-tetradeoxy-4-hydroxyformamido-3-C-methyl-3-nitro-d-hexose; 14, TcaA5-ACP-bound polyketide intermediate; 15, glyceroyl-TcaD2; 16, glyceroyl-CoA; 17, intermediate from the condensation of 14 and 16; 18, five-membered ring lactone-containing intermediate from the cyclization of 17; 19, intermediate from the dehydration of 18; 20, pretetronolide; 21, tetronolide.
TcaB1 may initiate the common steps to form these three sugar units by the conversion of d-glucose-1-phosphate (compound 1) to TDP-d-glucose (compound 2). TcaB2 is assumed to catalyze the formation of TDP-6-deoxy-4-keto-d-glucose (compound 3), a universal intermediate found in many biosynthetic pathways of deoxysugars. TcaB3 could be responsible for C-2 deoxygenation, yielding TDP-2,6-dideoxy-3,4-diketo-d-glucose (compound 4) as the first branch point between deoxysugar (l-amicetose and l-digitoxose) and deoxynitrosugar (d-tetronitrose) pathways.
TcaB4 shows high sequence homology to sugar 3-ketoreductases that reduce the C-3-keto group to form an equatorial hydroxyl group; therefore, its product is predicted to be TDP-2,6-dideoxy-4-keto-d-glucose (compound 5). TcaB5, belonging to a family of sugar 5-epimerases, may catalyze the conversion of compound 5 to TDP-2,6-dideoxy-4-keto-l-allose (compound 6), which serves as the second branch point between the l-amicetose and l-digitoxose pathways. The putative enzyme that acts on compound 6 to form TDP-2,3,6-trideoxy-4-keto-l-hexose (compound 7) appears unclear. It is most likely that an unknown sugar 3,4-dehydratase gene outside of the tca gene cluster performs this function. Next, reductive reactions for the introduction of the hydroxyl groups at the C-4 positions of compounds 6 and 7 are required to reach both l-digitoxose and l-amicetose. Although no apparent sugar 4-ketoreductase was found, TcaB6, showing high sequence homology to a NAD(P)H-dependent aldo/keto reductase family, could serve as a candidate to provide this broad substrate activity. Notably, kijA2 in the kij gene cluster encodes a TcaB6-homologous protein (28% identity) that shows sequence similarity to TylC1 for the 3-ketoreduction to form the C-3 epimer of compound 5 but was predicted not to be involved in KIJ deoxysugar biosynthesis (44). It would be interesting to determine whether TcaB6 acts on both compounds 6 and 7 to produce l-digitoxose and l-amicetose, respectively.
TcaB8, resembling members of a sugar 3-aminotransferase family, may act on compound 4 as a TcaB4 competitor to form TDP-2,3,6-trideoxy-4-keto-3-amino-d-glucose (compound 8) and switch the pathway to d-tetronitrose, one of the most highly functionalized sugars found in nature, as shown in Fig. 3A. TcaB9, with high sequence similarity to various sugar 3-methyltransferases, is assumed to be responsible for the conversion of compound 8 to TDP-2,3,6-trideoxy-4-keto-3-amino-3-C-methyl-d-hexose (compound 9). Similar to KijD3, TcaB10 shows high sequence homology to RubN8 (62% identity) in the biosynthesis of rubradirin (15), a nitrosugar-containing natural product, suggesting that TcaB10 acts on the C-3 amino group as an oxidase to render the rarely found nitro group, yielding compound 10. Subsequently, TcaB7 could serve as a sugar 4-aminotransferase to convert compound 10 to compound 11. TcaB11, homologous to an S-adenosylmethionine-dependent methyltransferase family, likely catalyzes the N-methylation on the C-4 amino group to form compound 12 as KijD8. However, no homologs of kijB3 (a flavin-dependent oxidoreductase gene) and kijA1 (an O-methyltransferase gene) were identified in the tca gene cluster for methylcarbamate moiety formation, implying that these functions might be encoded by unknown genes outside of this DNA region.
Genes involved in the biosynthesis of tetronolide.
Five genes, tcaA1 to tcaA5, were identified within the tca gene cluster, encoding the multifunctional modular type I PKSs, as shown in Fig. 3B. On the basis of sequence analysis, the first module of TcaA1 contains a mutant ketosynthase domain, in which a key active amino acid residue, cysteine, is replaced by a glutamine, supporting the idea that it functions as a loading module by decarboxylation of the ACP-tethered malonate unit to form an acetyl group and initiates the assembly process (16). To predict the substrates of the acyltransferase (AT) domains, several specificity-conferring residues in each were identified, showing the ATs of modules M4, M6, M7, and M11, specific to malonyl-CoA, and ATs of the remaining extending modules, as well as the loading module, specific to methylmalonyl-CoA (28). This is consistent with the structure of TCA on the whole; however, analysis of the TCA structure indicated that the ATs of the loading module, and modules M1 and M2, appear to be malonyl-CoA specific. Similar inconsistency was also found in the corresponding ATs of KIJ PKSs, supporting the notion that these three domains are exceptions to the current predictive model and that the PKS system of TCA may have an ancestor similar to that of KIJ for evolution (44). The dehydratase (DH) domain of module M8 in TcaA3 contains a mutant conserved motif in which the acidic residue glutamic acid is replaced by a neutral residue, alanine, indicating that this DH domain could be inactive (in accordance with the synthesized structure) (40). Notably, module M9 apparently lacks an enoylreductase domain to fully saturate the β-keto group, the activity of which is indispensable for folding of the latter intermediate to a conformation that facilitates a [4 + 2] cycloaddition to form the trans-decalin ring system. The upstream enoylreductase domain, which is exclusive to this PKS system and is functionally redundant in module M8 owing to the proposed nonfunctional cognate DH domain, therefore may carry this function. Sequence alignment revealed two inactive ketoreductase (KR) domains in modules M4 and M11, which apparently lack several conserved motifs characteristic of typical KRs (29, 40). According to a model for prediction of the stereochemistry of hydroxyacyl intermediates, the remaining nine KR domains were supposed to act on the keto groups to form d products exclusively (13). Consequently, TcaA1 to TcaA5, in a mechanistic analogy to ChlA1 to ChlA6, KijS1 to KijS5, and a number of typical type I PKSs, could catalyze the formation of a nascent tetracosanoyl-S-ACP intermediate via 11 decarboxylative condensations in a noniterative manner during the PKS-directed assembly process. The octahydronaphthalene ring structure of tetronate might be formed by an intramolecular cycloaddition via a “Diels-Alder”-like reaction, yielding intermediate 14.
Genetic comparison among the gene clusters of TCA, CHL, KIJ, and TMN led to the identification of four genes, tcaD1 to tcaD4, which were proposed to encode the characteristic tetronate moiety formation, as shown in Fig. 3B and 4A. Particularly in the chl, tca, and kij gene clusters, these genes constitute an independent operon and share head-to-tail similarity in both sequence and organization, supporting their highly functional association. TcaD1, similarly to ChlD1, KijC, and Tmn16, may carry both the AT and phosphatase activities to load the glyceroyl group from 1,3-BPG onto TcaD2, a discrete ACP protein homologous to ChlD2, KijD, and Tmn7a, yielding intermediate 15. This catalytic process was biochemically elucidated in vitro by the studies of ChlD1 and ChlD2/TcaD2 (described below) and of Tmn16 and Tmn7a (35), in a manner similar to OzmB and OzmE, which are involved in the utilization of the same substrate to form a relatively rare methyoxymalonyl unit for PKS-directed extension in oxazolomycin biosynthesis (6). TcaD3 appears to be a bifunctional protein like KijE, with the N-terminal domain homologous to ChlD3 and Tmn7 and the C-terminal domain homologous to ChlD4 and Tmn17. TcaD4, belonging to a ketosynthase III family, shows high sequence similarity to ChlM, KijB, and Tmn15. In our previous studies, gene inactivations of chlD3 and chlD4 completely abolished the production of CHL, confirming that they were essential to CHL biosynthesis; however, inactivation of chlM retained the ability to produce CHL, making its role in tetronate biosynthesis dubious (11). Using a set of CHL biosynthetic genes as probes, we recently identified two positive signals on the chromosome and an extrachromosomal plasmid upon pulsed-field gel electrophoresis analysis (unpublished data), suggesting that more than one putative (intact or not) chl gene cluster exist in S. antibioticus. This feature was also found in Streptomyces carzinostaticus, which harbors at least three gene clusters for neocarzinostatin biosynthesis (18). Thus, the function of chlM might not be completely inactivated by disruption of all putative copies in the analyzed mutants. Consequently, in a manner similar to the proposed functions of KijE and KijB, the N-terminal domain of TcaD3 may act as an AT to catalyze the transfer of the glyceroyl group from TcaD2 to CoA, forming glyceroyl-CoA, compound 16, which is then condensed with TcaA5-ACP-bound polyketide intermediate 14 to form compound 17 in the presence of TcaD4. Next, compound 17 could undergo an intramolecular cyclization catalyzed by the C-terminal domain of TcaD3 as a thioesterase, resulting in the five-member ring lactone functionality of intermediate 18.
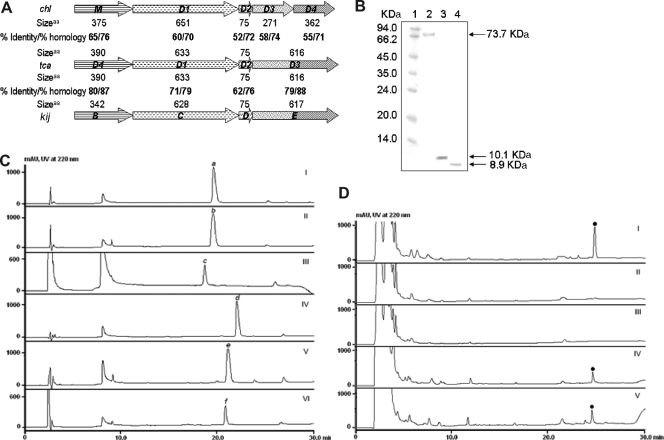
FIG. 4.
Investigation of tetronate biosynthesis. (A) Organization and sequence comparison of genes encoding tetronate biosynthesis for CHL, TCA, and KIJ. (B) Molecular mass marker (lane 1), purified ChlD1 (lane 2), ChlD2 (lane 3), and TcaD2 (lane 4) on tricine-polyacrylamide gel. (C) HPLC analysis of ChlD1-catalyzed loading of glyceroyl group onto ACPs. I, apo-ChlD2 (a), a negative control in the absence of Sfp; II, conversion of apo-ChlD2 to holo-ChlD2 (b) in the presence of Sfp; III, conversion of holo-ChlD2 to glyceroyl-ChlD2 (c) in the presence of ChlD1 within 10 min; IV, apo-TcaD2 (d), a negative control in the absence of Sfp; V, conversion of apo-TcaD2 to holo-TcaD2 (e) in the presence of Sfp; VI, conversion of holo-TcaD2 to glyceroyl-TcaD2 (f) in the presence of ChlD1 within 10 min. (D) HPLC analysis of CHL (solid dots) production from wild-type S. antibioticus and recombinant strains. I, wild type; II, TL1006 (ΔchlD3); III, TL1007 (ΔchlD4); IV, TL3001 (ΔchlD3/tcaD3); and V, TL3002 (ΔchlD4/tcaD3).
TcaE1, a putative flavin-dependent protein, shows high sequence similarity to KijA, which was proposed to catalyze the dehydration to form a tetronoate intermediate similar to compound 19. However, the possibility that this dehydration step occurs spontaneously to form a relatively stable conjugated system could not be excluded. The resulting double bond may promote an intramolecular Diels-Alder-like cycloaddition to close the macrocyclic ring and yield pretetronolide 20. According to this hypothesis, TcaE1 may function as an oxidase to form an aldehyde group at the C-23 position. Since no homologs of TcaE2 that resemble a flavin-dependent oxidoreductase family were found in the CHL and KIJ biosynthetic pathways, TcaE2 could act either as an oxidoreductase to form a hydroxyl group at the C-17 position for the tetronitrose unit appended to it or as a hydroxylase to introduce a regiospecific hydroxyl group at the C-21 position for maturation of the agylcone.
Genes associated with glycosyl transfer reactions and tailoring steps.
Four genes, tcaT1 to tcaT4, within the tca gene cluster were identified as encoding the glycosyltransferases (GTs), as shown in Fig. 2B. Sequence alignment revealed that these GTs show high sequence similarities to various GTs with microbial origins, in particular to ChlC7 (37 to 42% identity), supporting their actions on the tetronolide-like substrates as the sugar receptors. Intriguingly, since five deoxy(nitro)sugars exist in the structure of TCA and the GT for the d-tetronitrose attachment at the C-17 position should be unique, one GT might be involved twice in the formation of the tetrosaccharide side chain at the C-9 position. It is most likely the candidate for appending of two l-amicetoses, but with distinct glycosidic linkage patterns shown as α(1→3) and β(1→4), respectively. Similar to KIJ, the second digitoxose is attached via an α(1→4) linkage, indicating that this GT-specific glycosylation proceeds with retention of the configuration. It is not possible to predict their substrate specificities based upon the sequence analysis alone, and systematic inactivation of individual GT genes has been carried out to determine their action order.
One gene in the tca gene cluster, tcaM, encodes a protein similar to an O-AT family. TcaM could be responsible for the 4-O-acetylation on the first digitoxose that is attached to the aglycone via an α linkage. On the other hand, multiple glycosylations can begin with pretetronolide 20, and oxidations to form the hydroxyl and aldehyde functional groups therefore may occur as tailoring steps.
Characterization of the genes/proteins associated with tetronate biosynthesis.
Several attempts to determine the TcaD1 activity in vitro were hampered by poor expression of the resultant protein. Therefore, the homolog ChlD1 and the cognate ACP proteins ChlD2 and TcaD2 were heterologously expressed in E. coli, yielding the soluble proteins that were subsequently purified to near homogeneity as shown in Fig. 4B. Because the ACPs overproduced in E. coli are in the nonfunctional apo forms, incubation of ChlD2 and TcaD2 with CoA and Sfp led to their complete conversion into the functional holo forms, as judged by HPLC and MALDI-TOF MS analyses. To determine the activity of ChlD1, the purified protein was incubated with ATP, Mg2+, d-3-PG, TCEP, and PGK (for the generation of 1,3-BPG in situ), followed by addition of the reaction mixture containing holo-ChlD2 or holo-TcaD2. The resulting reaction mixtures were subjected to HPLC and MALDI-TOF MS analyses, revealing the complete conversion of holo-ChlD2 to glyceroyl-ChlD2 or holo-TcaD2 to glyceroyl-TcaD2 (Fig. 4C and Table 3; see Figure S1 in the supplemental material) within 10 min. Further incubation regenerated holo-ChlD2 or holo-TcaD2, probably due to the hydrolysis of glycerate from the ACP (data not shown). These findings indeed confirmed that ChlD1 bears both AT and phosphatase activities to load the glyceroyl group onto the ACP. Nearly identical conversion efficiencies under these reaction conditions were found by using either ChlD2 or TcaD2 as the ACP, indicating their extreme functional similarity. During our characterization work, similar results were obtained from more detailed studies of Tmn16 and Tmn7a in TMN biosynthesis (35).
TABLE 3.
MALDI-TOF MS analysis of apo-, holo-, and glyceroyl-ACPs
| ACP | Apo-ACP [M]+
|
Holo-ACP [M]+
|
Glyceroyl-ACP [M]+
|
|||
|---|---|---|---|---|---|---|
| Calculated | Found | Calculated | Found | Calculated | Found | |
| ChlD2 | 10,130.3 | 10,133.6 | 10,470.4 | 10,473.5 | 10,558.4 | 10,558.3 |
| TcaD2 | 8,951.2 | 8,950.0 | 9,290.9 | 9,291.0 | 9,378.9 | 9,378.1 |
In our previous study, inactivation of chlD3 and chlD4 resulted in two non-CHL-producing strains, TL1006 and TL1007, respectively. tcaD3, found in the tca gene cluster, is likely a fused gene that involves both the chlD3 and chlD4 functions upon sequence analysis. To verify this hypothesis, a construct that carried tcaD3 alone under the control of PermE* (the constitutive promoter of the erythromycin resistance gene eryE) (34) was introduced into TL1006 and TL1007, respectively, yielding the recombinant strain TL3001 (ΔchlD3/tcaD3) and TL3002 (ΔchlD4/tcaD3). As expected, HPLC analysis of the fermentation broths of both TL3001 and TL3002 revealed a compound with the same retention time as CHL (Fig. 4D), the identity of which was further confirmed by HPLC-MS analysis, strongly supporting the notion that TcaD3 can effectively replace the function of ChlD3 or ChlD4 and associate with the native proteins ChlD1, ChlD2, and ChlM to form the tetronate moiety for CHL production. Together, these findings clearly indicate that the tetronate-containing natural products share a highly conserved mechanism for this characteristic moiety biosynthesis.
Validation of the tca gene cluster.
TCA is produced by M. chalcea. Although the general methods well developed for various Streptomyces species could be applied to access the Micromonospora strains, the genetic system specific for the TCA-producing strain had not been established before our study. Since this is a nonsporulation strain, initial attempts were focused on the approach of polyethylene glycol-mediated protoplast transformation. However, extensive efforts failed due to the high resistance of the cell wall to lysozyme digestion and the poor regeneration efficiency of protoplasts. Eventually, using an intergeneric conjugation method with highly fragmented mycelia as the receptor cells (2, 23), the foreign DNAs were successfully introduced into the TCA-producing strain, establishing the premise for functional studies of the TCA biosynthetic genes in vivo.
To confirm the cloned gene cluster responsible for TCA biosynthesis, a large DNA fragment that spanned a 30,146-bp region containing partial tcaA1 and tcaA5 and intact tcaA2 and tcaA4 genes was deleted by double-crossover homologous exchange, yielding the mutant strain TL3003. With the wild-type strain as a control, the mutant strain indeed lost the ability to produce TCA, as shown in Fig. 5, unambiguously confirming that these PKS genes are essential to TCA production. Additionally, the disappearance of a number of other compounds in the mutant's extract suggested that they are structurally related to TCA, such as intermediates, by-products, or derivatives resulting from further biotransformation in the cells. For example, besides the TCA analogs found previously (e.g., TCB that lacks the terminal digitoxose moiety, with an [M+H]+ ion at an m/z of 1,199.08 and an [M+Na]+ ion at an m/z of 1,221.14), a few putative novel compounds that show glycosylation patterns identical (or similar) to those of TCA but contain changes on aglycones were identified (see Table S1 and Fig. S2 in the supplemental material) upon analysis of their quasimolecular and fragmentary ions. Elucidation of their structures will undoubtedly contribute to a better understanding of the TCA biosynthetic machinery. This result not only confirmed the cloned TCA biosynthetic gene cluster but also provided a clear background as the negative control to identify the novel TCA analogs from recombinant strains generated by genetic engineering of the TCA biosynthetic pathway.
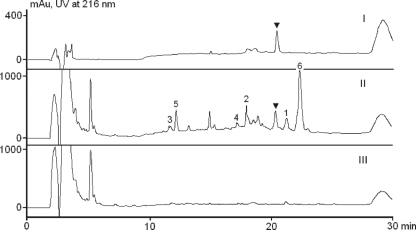
FIG. 5.
HPLC analysis of the production of TCA and related analogs from wild-type M. chalcea and recombinant strain TL3003. I, authentic standard; II, wild type; III, TL3003 (ΔtacA1, -A2, -A4, and -A5). The solid triangles indicate TCA. The numbers 1 to 6 indicate TCA analogs 1 to 6.
In conclusion, the availability of the tca gene cluster and the proposed biosynthetic pathway described here provide an excellent opportunity to access many interesting enzymatic mechanisms. The biosynthesis of unusual sugars (i.e., d-tetronitrose, l-amicetose, and l-digitoxose) may begin with the same precursor, d-glucose-1-phosphate; share early enzymatic steps toward common intermediates, TDP-2,6-dideoxy-3,4-diketo-d-glucose among all three and TDP-2,6-dideoxy-4-keto-l-allose between the last two; and branch into different pathways through competitive actions of specific enzymes. The biosynthesis of tetronolide might share a common strategy with those for chlorothricolide of CHL and kijanolide of KIJ, including the assembly of the aglycone skeleton by incorporation of a glycerol-derived 3-C unit with a polyketide intermediate and the involvement of two intramolecular cycloadditions to form the spirotetronate and trans-decarlin ring system. The resulting aglycone could be further equipped with five deoxy(nitro)sugars by employing four GTs to furnish the TCA structure. For tetronate formation, the first enzymatic step for transfer of a glyceroyl group onto a discrete ACP protein, ChlD2/TcaD2, was characterized in vitro, consistent with the functions of Tmn16 and Tmn7a found in TMN biosynthesis. Heterologous expression of the bifunction-fused gene tcaD3 in the non-CHL-producing mutants TL1006 (ΔchlD3, encoding a protein similar to the N terminus of TcaD3) and TL1007 (ΔchlD4, encoding a protein similar to the C terminus of TcaD3) restored CHL production by cross-complementation, strongly supporting the highly conserved strategy for tetronoate moiety biosynthesis in spirotetronate antibiotics. Finally, the biosynthetic machinery of TCA is amenable to genetic manipulation. The resultant pks deletion mutant serves as an excellent negative control for the identification of unknown related compounds from the wild-type strain and leading to novel analogs from recombinant strains generated by genetic engineering of the TCA biosynthetic pathway.
Supplementary Material
Acknowledgments
We thank the U.S. National Cancer Institute (NCI) for providing the TCA standard and Hung-Wen Liu, University of Texas at Austin, for proving the sequence of the kij gene cluster before its public release.
This work was supported in part by grants from the National Natural Science Foundation of China (30770035, 30525001, 90713012, 20321202, and 20402022), the Ministry of Science and Technology of China (2006AA020304 and 2006AA022185), the Chinese Academy of Science (KJCX2-YW-H08 and KJCX2-YW-G015), and the Science and Technology Commission of Shanghai Municipality (074319115-3, 05QMX1468, 05PJ14112, and 04DZ14901).
Footnotes
Published ahead of print on 27 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anether, G., I. Tinhofer, M. Senfter, and R. Greil. 2003. Tetrocarcin-A-induced ER stress mediates apoptosis in B-CLL cells via a Bcl-2-independent pathway. Blood 1014561-4568. [DOI] [PubMed] [Google Scholar]
- 2.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. Nagaraja, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 11643-49. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard, S., and J. S. Thorson. 2006. Enzymatic tools for engineering natural product glycosylation. Curr. Opin. Chem. Biol. 10263-271. [DOI] [PubMed] [Google Scholar]
- 4.Boeckman, R. K., P. Shao, S. T. Wrobleski, D. J. Boehmler, G. R. Heintzelman, and A. J. Barbosa. 2006. Toward the development of a general chiral auxiliary. A total synthesis of (+)-tetronolide via a tandem ketene-trapping [4 + 2] cycloaddition strategy. J. Am. Chem. Soc. 12810572-10588. [DOI] [PubMed] [Google Scholar]
- 5.Demydchuk, Y., Y. Sun, H. Hong, J. Staunton, J. B. Spencer, and P. F. Leadlay. 2008. Analysis of the tetronomycin gene cluster: insights into the biosynthesis of a polyether tetronate antibiotic. Chembiochem 91136-1145. [DOI] [PubMed] [Google Scholar]
- 6.Dorrestein, P. C., S. G. Van Lanen, W. Li, C. Zhao, Z. Deng, B. Shen, and N. L. Kelleher. 2006. The bifunctional glyceryl transferase/phosphatase OzmB belonging to the HAD superfamily that diverts 1,3-bisphosphoglycerate into polyketide biosynthesis. J. Am. Chem. Soc. 12810386-10387. [DOI] [PubMed] [Google Scholar]
- 7.Hallis, T. M., and H.-W. Liu. 1999. Learning nature's strategies for making deoxy sugars: pathways, mechanisms, and combinatorial applications. Acc. Chem. Res. 32579-588. [Google Scholar]
- 8.Hara, T., M. Omura-Minamisawa, C. Chao, Y. Nakagami, M. Ito, and T. Inoue. 2005. Bcl-2 inhibitors potentiate the cytotoxic effects of radiation in Bcl-2 overexpressing radioresistant tumor cells. Int. J. Radiat. Oncol. Biol. Phys. 61517-528. [DOI] [PubMed] [Google Scholar]
- 9.He, X. M., and H.-W. Liu. 2002. Formation of unusual sugars: mechanistic studies and biosynthetic applications. Annu. Rev. Biochem. 71701-754. [DOI] [PubMed] [Google Scholar]
- 10.Holzbach, R., H. Page, D. Hook, E. F. Kreuzer, C. Chang, and H. G. Floss. 1978. Biosynthesis of the macrolide antibiotic chlorothricin: basic building blocks. Biochemistry 17556-560. [DOI] [PubMed] [Google Scholar]
- 11.Jia, X., Z. Tian, L. Shao, X. Qu, Q. Zhao, J. Tang, G. Tang, and W. Liu. 2006. Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem. Biol. 13575-585. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko, M., T. Nakashima, Y. Uosaki, M. Hara, S. Ikeda, and Y. Kanda. 2001. Synthesis of tetrocarcin derivatives with specific inhibitory activity towards Bcl-2 functions. Bioorg. Med. Chem. Lett. 11887-890. [DOI] [PubMed] [Google Scholar]
- 13.Keatinge-Clay, A. T. 2007. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 14898-908. [DOI] [PubMed] [Google Scholar]
- 14.Kieser, T., M. Bibb, M. Butter, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 15.Kim, C. G., J. Lamichhane, K. I. Song, V. D. Nguyen, D. H. Kim, T. S. Jeong, S. H. Kang, K. W. Kim, J. Maharjan, Y. S. Hong, J. S. Kang, J. C. Yoo, J. J. Lee, T. J. Oh, K. Liou, and J. K. Sohng. 2007. Biosynthesis of rubradirin as an ansamycin antibiotic from Streptomyces achromogenes var. rubradiris NRRL3061. Arch. Microbiol. doi: 10.1007/s00203-007-0337-3. [DOI] [PubMed]
- 16.Kim, E., K. D. Cramer, A. L. Shreve, and D. H. Sherman. 1995. Heterologous expression of an engineered biosynthetic pathway: functional dissection of type II polyketide synthase components in Streptomyces species. J. Bacteriol. 1771202-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, J. J., J. P. Lee, P. J. Keller, C. E. Cottrell, C. Chang, H. Zähner, and H. G. Floss. 1986. Further studies on the biosynthesis of chlorothricin. J. Antibiot. 391123-1134. [DOI] [PubMed] [Google Scholar]
- 18.Liu, W., K. Nonaka, L. Nie, J. Zhang, S. D. Christenson, J. Bae, S. G. Van Lanen, E. Zazopoulos, C. M. Farnet, C. F. Yang, and B. Shen. 2005. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem. Biol. 12293-302. [DOI] [PubMed] [Google Scholar]
- 19.Liu, W., and B. Shen. 2000. Genes for production of the enediyne antitumor antibiotic C-1027 in Streptomyces globisporus are clustered with the cagA gene that encodes the C-1027 apoprotein. Antimicrob. Agents Chemother. 44382-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luzhetskyy, A., and A. Bechthold. 2005. It works: combinatorial biosynthesis for generating novel glycosylated compounds. Mol. Microbiol. 583-5. [DOI] [PubMed] [Google Scholar]
- 21.Mallams, A. K., M. S. Puar, and R. R. Rossman. 1981. Kijanimicin. 1. Structures of individual sugar components. J. Am. Chem. Soc. 1033938-3940. [Google Scholar]
- 22.Mascaretti, O. A., C. Chang, D. Hook, H. Otsuka, E. F. Kreuzer, and H. G. Floss. 1981. Biosynthesis of the macrolide antibiotic chlorothricin. Biochemistry 20919-924. [DOI] [PubMed] [Google Scholar]
- 23.Mazodier, H., R. Petter, and C. Thompson. 1989. Intergeneric conjugation between Escherichia and Streptomyces species. J. Bacteriol. 1713583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto, M., M. Fukui, S. Ohkubo, T. Tamaoki, and F. Tomita. 1982. Tetrocarcins, new antitumor antibiotics. 3. Antitumor activity of tetrocarcin A. J. Antibiot. 351033-1037. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima, H., K. Sakaguchi, I. Fujiwara, M. Mizuta, M. Tsuruga, J. Magae, and N. Mizuta. 2007. Apoptosis and inactivation of the PI3-kinase pathway by tetrocarcin A in breast cancers. Biochem. Biophys. Res. Commun. 356260-265. [DOI] [PubMed] [Google Scholar]
- 26.Nakashima, T., M. Miura, and M. Hara. 2000. Tetrocarcin A inhibits mitochondrial functions of Bcl-2 and suppresses its anti-apoptotic activity. Cancer Res. 601229-1235. [PubMed] [Google Scholar]
- 27.Quadri, L. E. N., P. H. Weinreb, M. Lei, M. M. Nakano, P. Zuber, and C. T. Walsh. 1998. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 371585-1595. [DOI] [PubMed] [Google Scholar]
- 28.Reeves, C. D., S. Murli, G. W. Ashley, M. Piagentini, R. C. Hutchinson, and R. McDaniel. 2001. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry 4015464-15470. [DOI] [PubMed] [Google Scholar]
- 29.Reid, R., M. Piagentini, E. Rodriguez, G. Ashley, N. Viswanathan, J. Carney, D. V. Santi, C. R. Hutchinson, and R. McDaniel. 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 4272-79. [DOI] [PubMed] [Google Scholar]
- 30.Roush, W. R., and R. Sciotti. 1994. Enantioselective total synthesis of (−)-chlorothricolide. J. Am. Chem. Soc. 1166457-6458. [Google Scholar]
- 31.Roush, W. R., and R. Sciotti. 1998. Enantioselective total synthesis of (−)-chlorothricolide via tandem inter- and intra-molecular Diels-Alder reaction of a hexaenoate intermediate. J. Am. Chem. Soc. 1207411-7419. [Google Scholar]
- 32.Salas, J. A., and C. Mendez. 2007. Engineering the glycosylation of natural products in actinomycetes. Trends Microbiol. 15219-232. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schmitt-John, T., and J. W. Engels. 1992. Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl. Microbiol. Biotechnol. 36493-498. [DOI] [PubMed] [Google Scholar]
- 35.Sun, Y., H. Hong, F. Gillies, J. B. Spencer, and P. F. Leadlay. 2008. Glyceryl-S-acyl carrier protein as an intermediate in the biosynthesis of tetronate antibiotics. Chembiochem 9150-156. [DOI] [PubMed] [Google Scholar]
- 36.Takeda, K., E. Kawanishi, H. Nakamira, and E. Yoshii. 1991. Total synthesis of tetronolide, the aglycon of tetrocarcins. Tetrahedron Lett. 324925-4928. [Google Scholar]
- 37.Tamaoki, T., M. Kasai, K. Shirahata, S. Ohkubo, M. Morimoto, K. Mineura, S. Ishii, and F. Tomita. 1980. Tetrocarcins, novel antitumor antibiotics. II. Isolation, characterization and antitumor activity. J. Antibiot. 33946-950. [DOI] [PubMed] [Google Scholar]
- 38.Tamaoki, T., and F. Tomita. 1982. Biosynthesis and production of tetrocarcin A, a new antitumor antibiotic in chemically defined medium. Agric. Bio. Chem. 461021-1026. [Google Scholar]
- 39.Tamaoki, T., and F. Tomita. 1983. Biosynthesis of tetrocarcin. Incorporation of 14C- and 13C-labeled compounds into tetrocarcin. J. Antibiot. 36595-598. [DOI] [PubMed] [Google Scholar]
- 40.Tang, L., Y. J. Yoon, C. Choi, and C. R. Hutchinson. 1998. Characterization of the enzymatic domains in the modular polyketide synthase involved in rifamycin B biosynthesis by Amycolatopsis mediterranei. Gene 216255-265. [DOI] [PubMed] [Google Scholar]
- 41.Tinhofer, I., G. Anether, M. Senfter, K. Pfaller, D. Bernhard, M. Hara, and R. Greil. 2002. Stressful death of T-ALL tumor cells after treatment with the anti-tumor agent tetrocarcin-A. FASEB J. 161295-1297. [DOI] [PubMed] [Google Scholar]
- 42.Tomita, F., T. Tamaoki, K. Shirahata, M. Kasai, M. Morimoto, S. Ohkubo, K. Mineura, and S. Ishii. 1980. Novel antitumor antibiotics, tetrocarcins. J. Antibiot. 33668-670. [DOI] [PubMed] [Google Scholar]
- 43.Tomita, F., and T. Tamaoki. 1980. Tetrocarcins, novel antitumor antibiotics. I. Producing organism, fermentation and antimicrobial activity. J. Antibiot. 33940-945. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, H., J. A. White-Phillip, C. E. Melançon III, H. J. Kwon, W. L. Yu, and H.-W. Liu. 2007. Elucidation of the kijanimicin gene cluster: insights into the biosynthesis of spirotetronate antibiotics and nitrosugars. J. Am. Chem. Soc. 12914670-14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.