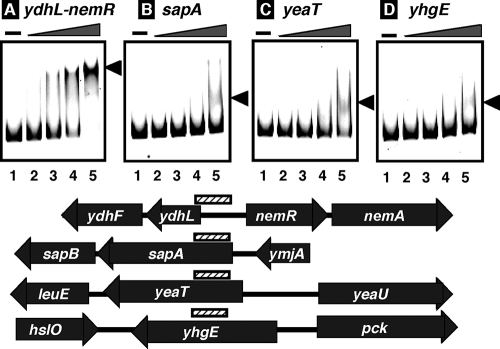
FIG. 1.
Gel mobility shift assay of NemR-target DNA complex formation. Fluorescence-labeled DNA probes, 1.0 pmol each, containing a SELEX segment from the ydhL-nemR spacer (A), sapA coding frame (B), yeaT coding frame (C), and yhgE coding frame (D), were incubated at 37°C for 30 min in the absence (lanes 1) or presence of increasing concentrations of NemR (lanes 2, 0.25 pmol; lanes 3, 0.5 pmol; lanes 4, 1.0 pmol; lanes 5, 2.0 pmol). The reaction mixtures were directly subjected to PAGE. The gene organization and the location of probes along the E. coli genome are shown below the gel patterns. The functions of known genes are as follows: nemA, NEM reductase; sapA/sapB, antimicrobial peptide transporter subunits; leuE, neutral amino acid efflux system; hslO, heat shock protein Hsp33; pck, phosphoenolpyruvate carboxykinase.