FIG. 1.
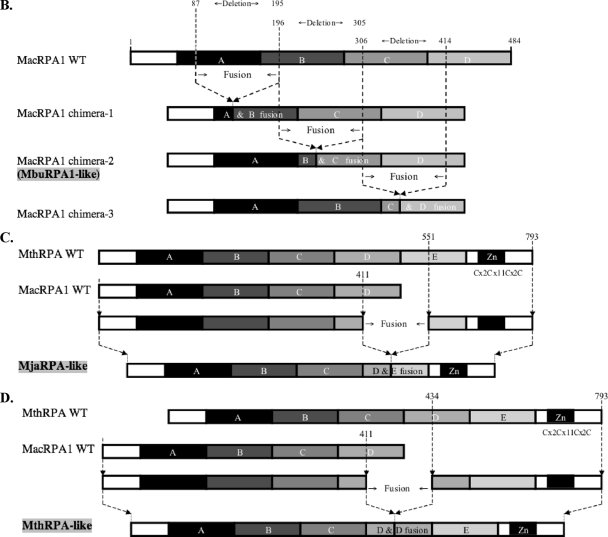
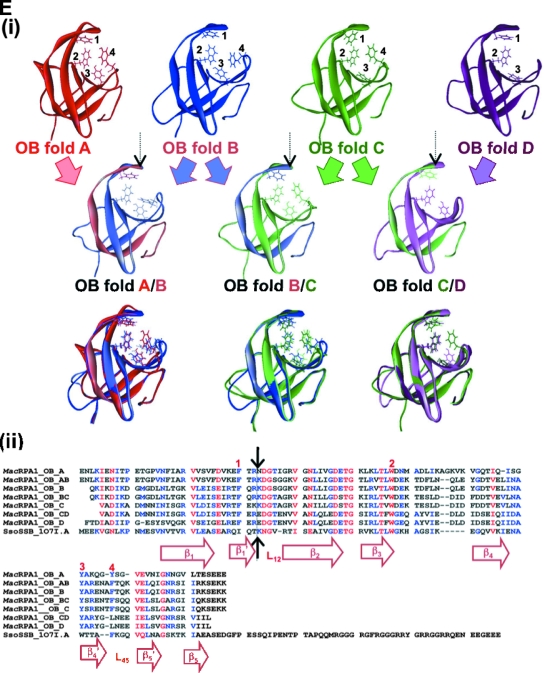
Alignment, schematic representation of MacRPA1 and related RPA or RPA-like proteins, and homology modeling of naturally occurring and artificially synthesized OB folds. (A) Alignment of MacRPA1 and related proteins. The proteins are M. acetivorans RPA1 (MacRPA1) (GenBank accession no. AAM07979), M. mazei RPA1 (MmaRPA1) (GenBank accession no. NP_633323), M. barkeri RPA1 (MbaRPA1) (GenBank accession no. ZP_00542400), Methanococcoides burtonii RPA1-like protein (MbuRPA1) (GenBank accession no. ZP_00563288), Methanosaeta thermophila RPA1-like protein (MtheRPA) (GenBank accession no. YP_843314), Haloarcula marismortui RPA1-like protein (HmaRPA) (GenBank accession no. AAV47126), Halobacterium sp. strain NRC-1 RPA1-like protein (HspRPA) (GenBank accession no. NP_279274), and Natronomonas pharaonis RPA1-like protein (NphRPA) (GenBank accession no. YP_325819). The amino acid sequences for the different OB folds in MacRPA1 are delineated with the differently shaded lines and are labeled with the letters A, B, C, and D. Identical amino acids (white letters on a black background), conserved amino acids (black letters on gray background), and gaps introduced to maximize sequence alignment (dashes) are indicated. (B) Schematic representations showing the construction of MacRPA1 chimeras (MacRPA1 chimera-1, MacRPA1 chimera-2, and MacRPA1 chimera-3) from wild-type (WT) MacRPA1. The schematic representations of the MacRPA1 chimeras show OB folds A to D. (C) Schematic representations showing the construction of M. jannaschii RPA-like protein (MjaRPA-like protein). The first three OB folds (folds A to C) and the N-terminal third of the fourth OB fold of M. acetivorans RPA1 were fused to the indicated modules from M. thermautotrophicus RPA through fusion of adjacent OB folds. (D) Schematic representations showing the construction of MthRPA-like protein. The same fragment from wild-type MacRPA1 for construction of the MjaRPA-like protein as shown in panel B was used, but the fusion fragment from wild-type (WT) MthRPA was from the fourth OB fold (instead of the fifth) to the end of the polypeptide. The full-length and chimeric (fused) OB folds are shown by differently shaded boxes with letters A, B, C, D, and E. The boxes denoted by Zn represent zinc-binding domains with their sequences (x represents any amino acid residue). The motifs are not drawn to scale, and the exact points of fusion are described in Materials and Methods. (E) The highly conserved structures of archaeal OB folds allow for functional fusions. (i) The top row shows homology models for the four MacRPA1 OB folds. The four aromatic residues (depicted by stick representation) expected to form critical interactions with ssDNA were identified on the basis of the comparison of amino acid sequence alignment and structural alignment between MacRPA1 OB folds and that of the SsoSSB. Note that OB fold D contains a leucine in the position occupied by tyrosine or phenylalanine in most known OB folds. The number by each residue identifies it in the alignment shown in panel ii. The middle row shows fusion OB folds. The elements of each fusion fold corresponding to the original OB folds are shown in the faded parental colors. The site of the fusion is indicated by the arrow. The bottom row shows superimposition of the parental and fusion folds. (ii) Amino acid sequence alignment of individual MacRPA1 OB folds (MacRPA1_OB_A to MacRPA1_OB_D), chimeric OB folds (MacRPA1_OB_AB to MacRPA1_OB_CD), and SsoSSB protein (SsoSSB_1O7I.A). Numbers above the alignment indicate residues proposed to interact with ssDNA. Black arrows indicate the position of the fusion. Elements of the secondary structure are indicated under the alignment. Sequences were aligned using Muitalin 5.4.1 (4) by comparison with a table constructed using the Blosum62 substitution matrix with a gap opening weight of 7 and gap extension weight of 1.