Figure 3. Comparison of Hetero- and Homophilic Interactions by SLAM Family Receptors.
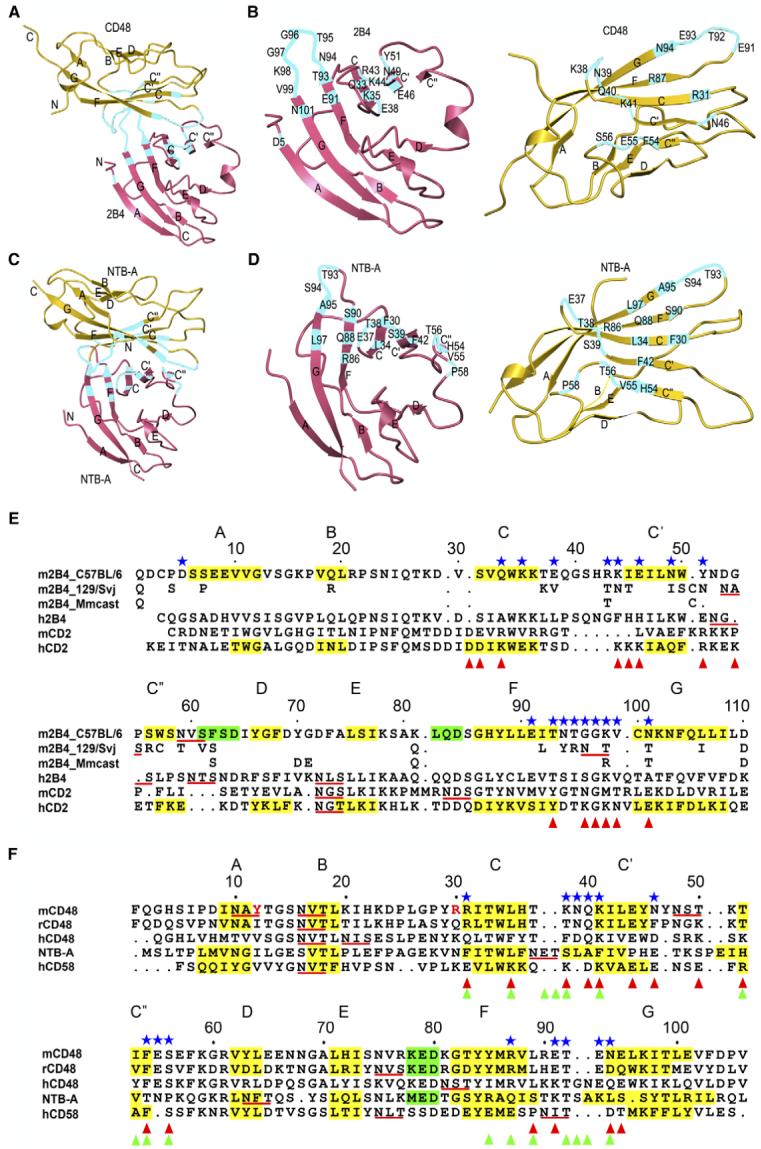
(A and C) Overall arrangement of the 2B4-CD48 heterophilic and NTB-A homophilic dimers. CD48 and the upper NTB-A monomer are in the same orientation. Residues contributing to the binding interfaces are highlighted in cyan. The N and C termini are indicated.
(B and D) Detailed view of the interfaces. CD48 and the upper NTB-A monomer are each rotated ∼90° about the horizontal axis relative to (A) and (C), respectively. Contacting residues are labeled.
(E and F) Structure-based alignment of CD2 family IgV domains. The β strands are shaded yellow and labeled A through G; α helices are shaded green. Potential N-linked glycosylation sites are underlined red. Contacting residues in the 2B4-CD48 heterophilic dimer are denoted with blue stars. Residues involved in homophilic interactions in the NTB-A dimer are marked with green triangles (Cao et al., 2006). Residues contributing to the CD2-CD58 heterophilic interface are denoted with red triangles (Wang et al., 1999). In (E), residues of 2B4 isoforms 129/Svj (UniProtKB-TrEMBL: Q3TDI8) and Mmcast (Mus musculus castaneus, UniProtKB-TrEMBL: Q3S4B1) that are identical to 2B4 isoform C57BL/6 (UniProtKB-Swiss-Prot: Q07763) residues have been omitted. In (F), residues of yeast-displayed mouse CD48 that differ from the wild-type sequence (Thr12, Lys30) are indicated in red.
