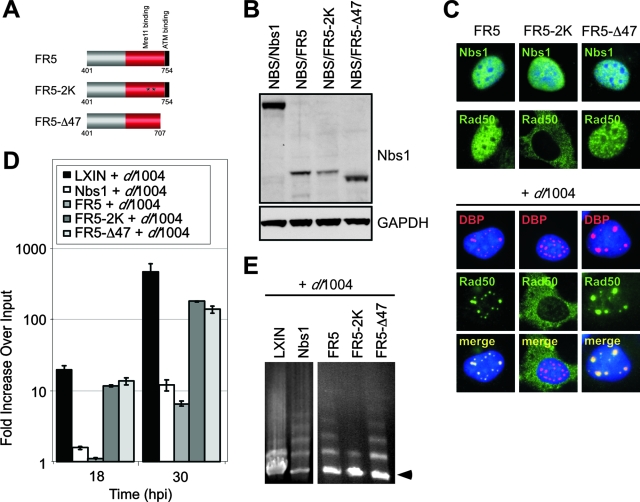
FIG. 7.
Requirements for inhibition of viral DNA replication and for concatemer formation by the C terminus of Nbs1. (A) Schematic of motifs in the C terminus of Nbs1 proteins and the mutations generated. (B) Immunoblot of lysates from NBS cells transduced with retroviruses expressing wild-type and mutant Nbs1 proteins. GAPDH served as a loading control. (C) Localization of Nbs1 and Rad50 proteins in NBS cells in the absence and presence of virus infection. NBS-derived cell lines either uninfected or infected with dl1004 were fixed and analyzed by immunofluorescence at 24 hpi. Virus replication centers were visualized in infected cells with an antibody to DBP, and cell nuclei were marked by staining with DAPI. (D) Effect of mutations in the C terminus FR5 fragment of Nbs1 on mutant Ad DNA replication. NBS cells expressing wild-type or mutant Nbs1 proteins were infected with mutant virus dl1004, and DNA was extracted over a time course for quantitation by qPCR. Accumulation of viral DNA is represented as the fold increase over input viral DNA, determined at the 4-h time point. (E) Complementation of concatemer formation by C-terminal fragments of Nbs1. NBS cells expressing FR5, FR5-2K and FR5-Δ47 were infected with dl1004, and DNA was prepared at 30 hpi for analysis by PFGE. Viral DNA was visualized by staining the gel in ethidium bromide, and the position of linear viral genome is indicated by an arrow.