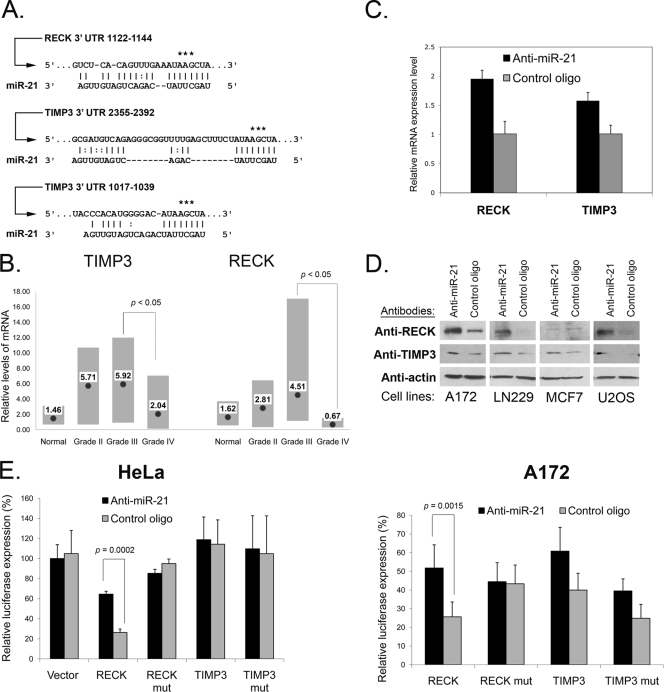
FIG. 4.
miR-21 targets RECK and TIMP3. (A) Predicted miR-21 binding sites within RECK and TIMP3 3′ UTRs. The asterisks depict nucleotides mutated for the luciferase reporter assays. (B) RECK and TIMP3 mRNA expression in normal brain tissue and gliomas of different grades was analyzed by qRT-PCR and normalized to GAPDH mRNA levels. Per group, four control brains and grade II gliomas and eight grade III and grade IV gliomas were analyzed. The gray bars represent the data ranges. Mean expression levels are depicted by black dots and corresponding numbers. Since a few samples were analyzed in “normal” and “grade II” sets, the analysis reaches significance (P < 0.05) for the transition from grade III to IV only. (C) RECK and TIMP3 mRNAs are upregulated upon treatment of glioma cells with anti-miR-21. A172 cells were transfected with either the anti-miR-21 or the control oligonucleotide (control oligo) and grown for 48 h, mRNA was isolated, and qRT-PCR for RECK and TIMP3 was performed. The data were normalized to GAPDH mRNA levels. Data are represented as means ± SEM. (D) Western blot validation of RECK and TIMP3 protein upregulation by anti-miR-21 in glioma (A172 and LN229) and other (MCF7, breast carcinoma; U2OS, osteosarcoma) cell lines. The analysis was performed at 72 h posttransfection. (E) pMir-Report vectors containing the wild-type RECK 3′ UTR (RECK wt), RECK 3′ UTR with a mutated miR-21 binding site (RECK mut), the wild-type TIMP3 3′ UTR (TIMP3 wt), or the TIMP3 3′ UTR with both predicted miR-21 binding sites mutated (TIMP3 mut) were cotransfected with either miR-21 inhibitor (Anti-miR-21) or the mismatched control oligonucleotide into A172 and HeLa cells. The inhibition of miR-21 by the antisense inhibitors resulted in a significant increase in luciferase signals of RECK wt- but not RECK mut-transfected cells. No consistent effect of miR-21 inhibition on the TIMP3 3′ UTR was observed. Depicted are the averages of results from representative experiments performed in triplicate. Data are represented as means ± SEM.