Abstract
Tubulin undergoes glutamylation, a conserved posttranslational modification of poorly understood function. We show here that in the ciliate Tetrahymena, most of the microtubule arrays contain glutamylated tubulin. However, the length of the polyglutamyl side chain is spatially regulated, with the longest side chains present on ciliary and basal body microtubules. We focused our efforts on the function of glutamylation on the α-tubulin subunit. By site-directed mutagenesis, we show that all six glutamates of the C-terminal tail domain of α-tubulin that provide potential sites for glutamylation are not essential but are needed for normal rates of cell multiplication and cilium-based functions (phagocytosis and cell motility). By comparative phylogeny and biochemical assays, we identify two conserved tubulin tyrosine ligase (TTL) domain proteins, Ttll1p and Ttll9p, as α-tubulin-preferring glutamyl ligase enzymes. In an in vitro microtubule glutamylation assay, Ttll1p showed a chain-initiating activity while Ttll9p had primarily a chain-elongating activity. GFP-Ttll1p localized mainly to basal bodies, while GFP-Ttll9p localized to cilia. Disruption of the TTLL1 and TTLL9 genes decreased the rates of cell multiplication and phagocytosis. Cells lacking both genes had fewer cortical microtubules and showed defects in the maturation of basal bodies. We conclude that glutamylation on α-tubulin is not essential but is required for efficiency of assembly and function of a subset of microtubule-based organelles. Furthermore, the spatial restriction of modifying enzymes appears to be a major mechanism that drives differential glutamylation at the subcellular level.
The principal components of microtubules, heterodimers of α- and β-tubulin, are known to undergo several types of conserved posttranslational modifications (PTMs), including acetylation, detyrosination, phosphorylation, palmitoylation, glutamylation, and glycylation (61). Some of these PTMs strongly influence interactions between microtubules and microtubule-associated proteins, including motors and plus-end tracking proteins (27, 32, 34, 43, 49). Specific PTMs are enriched on microtubules in restricted subcellular areas, suggesting that these mechanisms act as marks that locally adapt the microtubule polymer for specific functions (reviewed in reference 58). How the spatially restricted modified microtubules are generated within the cell is not well understood.
Glutamylated microtubules are generated by the sequential addition of multiple glutamates to the γ-carboxyl group of specific glutamic acids of the primary sequence of the C-terminal tail (CTT) domain of α- and β-tubulin (14). This reversible PTM creates glutamyl side chains of variable length and is enriched on microtubules in cilia, centrioles/basal bodies, the mitotic spindle, microtubules of nerve projections, and pellicular arrays in protists (2, 5, 6, 31, 35, 45, 63). Recently, we have identified tubulin glutamylases as proteins with a tubulin tyrosine ligase homology (28). The identification of forward enzymes for glutamylation opens the way for studies on the in vivo role of tubulin glutamylation and the mechanisms of spatial regulation of this PTM.
The ciliate Tetrahymena thermophila assembles 18 distinct types of microtubules within a single cell. Despite its structural complexity, the ease of reverse genetic methods has made Tetrahymena a useful model for studies on microtubule PTMs (20, 28, 48, 49, 55, 56, 64). Here we analyzed the cellular localization, in vivo function, and enzymatic activities of Tetrahymena homologs of two evolutionarily conserved glutamylase enzymes: tubulin tyrosine ligase (TTL)-like (TTLL) proteins TTLL1 and TTLL9. Both enzymes are α-tubulin-specific glutamylases. We complemented the analysis of TTLL1 and TTLL9 functions by creating a mutant that lacks sites of polyglutamylation on α-tubulin. We show that while glutamylation on α-tubulin is dispensable, it is required for the normal assembly and functions of a subset of microtubular structures within the cell. Furthermore, we show that the two enzymes that glutamylate α-tubulin each colocalize with a specific subset of microtubule types within the cell.
MATERIALS AND METHODS
Phylogenetic analysis of TTLL proteins.
Sequences of TTLL proteins were obtained from National Center for Biotechnology Information databases and the Tetrahymena Gene Database. For the gene accession numbers, see the legend to Fig. S1 in the supplemental material. For phylogenetic analysis, sequences were aligned with ClustalX 1.82 (29) and corrected manually in SEAVIEW (23). A tree was calculated with the Phylip package (SEQBOOT, PROTDIST, NEIGHBOR, CONSENSE, and DRAWGRAM) (16) by using 1,000 replicates and a Jones-Taylor-Thornton substitution model.
GFP tagging in vivo.
Cells were grown in SPP medium (24). The strain of T. thermophila expressing green fluorescent protein GFP-Ttll1p was described previously (28). We used a walking-primer reverse transcription-PCR to map the 5′ end of the mRNA encoding Ttll9p. The coding region of TTLL9, followed by 0.4 kb of the 3′ untranslated region (UTR), was amplified from genomic DNA with the addition of MluI and BamHI sites by using primers 5′-TTTATACGCGTCATGTCAGAATATTTTAGGAAATAC-3′ and 5′-ATTAAGGATCCGTTAGAGGTAAAGAAAGAAGA-3′, digested with MluI and BamHI, and ligated into plasmid pMTT1-GFP (62). The resulting pMTT1-GFP-TTLL9 plasmid was digested with SacII and ApaI and introduced into the CU522 strain of Tetrahymena by biolistic bombardment and selection with 20 μM paclitaxel (21). To induce the expression of GFP-tagged TTLLs, transgenic cells were grown overnight to a concentration of 2 × 105 to 3 × 105/ml without paclitaxel and incubated with CdCl2 (2.5 μg/ml) for 3 to 4 h.
Construction of strains lacking glutamylase genes or polymodification sites on α-tubulin.
The TTLL1 knockout strain of T. thermophila was constructed earlier (28). To prepare a plasmid for disruption of TTLL9, 2.5 kb of the 5′ UTR was amplified from genomic DNA with the addition of a SacII site (primer 5′-TTTTACCGCGGATTGTTCTTATTTTGGCTGGTT-3′) and a BamHI site (primer 5′-AATAAGGATCCGACAACGAACGGATAATGAATA-3′) at the 5′ and 3′ ends, respectively, and 2 kb of the coding region and the 3′ UTR was amplified with the addition of a SalI site (5′-ATAAAGTCGACGCAGAGCAATAATAATAGCAAT-3′) and an ApaI site (5′-TAAAAGGGCCCGAAATCAACTAAAATAAGAAGC-3′) at the 5′ and 3′ ends, respectively. The 2-kb fragment was cloned into plasmid p4T2ΔHindIII carrying the neo2 cassette by using SalI and ApaI sites. The resulting plasmid was digested with SacII and BamHI and used to clone the 2.5-kb fragment. The pTTLL9-neo2 plasmid was digested with SacII and ApaI and transformed biolistically into mating CU428 and B2086 cells (9). A ttll9::neo2 knockout heterokaryon (25) strain was produced. To make a double-knockout strain, a ttll1::neo2/ttll1::neo2 heterokaryon (28) was crossed to a ttll9::neo2/tll9::neo2 heterokaryon, double-heterozygous progeny cells were isolated and grown to enable loss of the neo2 alleles from the macronucleus by phenotypic assortment, and the micronucleus was made homozygous by a cross to B*VII (1). The clones homozygous for both disrupted loci in the micronucleus were identified by genotyping individual progeny of an outcross by using genomic PCR and pairs of primers that amplified a junction between neo2 and the flanking region of either TTLL1 or TTLL9. To obtain cells lacking both genes in the macronucleus, the double-knockout homozygous heterokaryons were crossed to each other and paromomycin-resistant progeny cells were recovered. The absence of TTLL1 and TTLL9 coding regions in the progeny cells was confirmed by PCR of total genomic DNA with primers designed to amplify the entire coding sequences (see Fig. S3 in the supplemental material).
To construct a strain of Tetrahymena that expresses an α-tubulin which cannot be polymodified in vivo, we used site-directed mutagenesis (33) to produce a plasmid containing the ATU1 gene fragment with mutations of all six glutamic acid codons to aspartic acid codons within the region encoding the CTT domain. The pATU1-6D fragment was used to rescue mating ATU1 knockout heterokaryons (26) to produce cells exclusively expressing the Atu1p-6D α-tubulin.
Microscopy and evaluation of phenotypes.
Tetrahymena cells were stained by immunofluorescence (62) with glutamylated-tubulin epitope-specific primary monoclonal antibodies (MAbs) GT335 (1:100) (63), B3 (1:1,000; Sigma Chemical Co. St. Louis, MO) (22), and ID5 (1:30) (53) and with polyE (1:100), a polyclonal antibody generated against a Cys-E9 peptide (54). Cells were viewed in a Leica TCS SP confocal microscope. For transmission electron microscopy, cells were prepared as described previously (30). To assay phagocytosis, cells were fed with 0.2% India ink in SPP for 2 to 10 min, fixed by adding an equal volume of 2% paraformaldehyde, and scored for the presence of labeled food vacuoles. To measure the rate of motility, we recorded the paths of motile cells on a Nikon TMS microscope (10× objective) with ImageJ 1.37 software and a DAGE MTI (Michigan City, IN) DC330 camera connected to a Scion (Scion Corporation, Frederick, MD) CG-7 frame grabber run on a Macintosh G4 computer. Straight paths recorded on five consecutive frames were measured with NIH Image 1.62. To measure growth rates, cells from a log-phase culture were diluted to 2 × 104/ml and grown without shaking at 30°C in 10 ml of SPP medium in a 150-ml flask. In some growth experiments, paclitaxel (LC Laboratories, Woburn, MA) or oryzalin (Chem Service, West Chester, PA) was used.
Biochemical procedures.
Cytoskeletons were extracted as described previously (28). To purify cilia, we modified the pH shock method (36) as follows. Cells grown in SPP (150 ml, 3 × 105/ml) were washed with Tris-HCl (pH 7.5) and suspended in 20 ml of 10 mM Tris-HCl (pH 7.4)-10 mM CaCl2-50 mM sucrose with protease inhibitors (12) in a 50-ml conical tube. Next, 350 μl of 0.5 M acetic acid was added (pH decreased to ∼4.3) and the cells were mixed by inverting the tube four times and incubated for 40 s, followed by the addition of 300 μl of 0.6 M KOH (pH increased to ∼7.5). Cell bodies were collected by centrifugation at 1,860 × g for 5 min, and cilia were recovered by centrifugation at 23,300 × g for 15 min. The ciliary pellet was suspended in 100 μl of PME buffer [100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 2.5 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 1 μM paclitaxel (pH 6.8)] with protease inhibitors and stored at −80°C. Ten micrograms of cytoskeletal protein or 5 μg of cilium protein per lane was separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis minigel under conditions in which Tetrahymena α-tubulin migrates faster than β-tubulin (48) and transferred to nitrocellulose. Western blot assays were done as described previously (28), with the ECL Advance Western blotting detection kit (GE Healthcare, Buckinghamshire, United Kingdom) and the primary antibodies GT335 (1:1,000), ID5 (1:50), polyE (1:1,000), 12G10 (anti-α-tubulin; 1:10,000; Developmental Studies Hybridoma Bank), polyG (anti-polyglycine; 1:5,000) (54), and 6-11-B1 (anti-acetylated K40 on α-tubulin; 1:10,000; Sigma Chemical Co., St. Louis, MO) and anti-GFP antibodies (1:2,000; Torrey Pines Biolabs, Houston, TX).
To partially purify either GFP-Ttll9p or GFP-Ttll1p, cells carrying MTT1 promoter-driven transgenes (150 ml, 3 × 105 cells/ml) were treated with 2.5 μg/ml CdCl2, washed with 10 mM Tris-HCl (pH 7.5), and lysed on ice in 3 ml of 50 mM Tris-HCl (pH 8.0), 0.2 M NaCl, 1 mM MgCl2, 1 mM EGTA, and 0.2% NP-40 with protease inhibitors. The cell lysate was centrifuged at 25,000 × g, the supernatant was loaded onto a phosphocellulose column, and bound proteins were eluted with a 0.2 to 1 M NaCl gradient. All fractions reactive with anti-GFP antibodies by Western blotting (0.5 to 0.6 M NaCl) were pooled, dialyzed, and concentrated to 1.5 mg/ml.
To measure tubulin glutamylase activity, the partially purified protein (0.15 mg/ml) was incubated with 50 mM Tris-HCl (pH 9.0), 0.4 mM ATP, 2.4 mM MgCl2, 0.5 mM dithiothreitol, 8 μM l-[3H]glutamate (45 to 55 Ci/mmol; GE Healthcare), and 0.1 mg/ml docetaxel (Taxotere)-stabilized microtubules from either HeLa cells or murine brains (51) at 30°C for 45 min. Fluorographic detection was performed by exposing a dried Coomassie blue-stained gel (enhanced in Amplify) to Hyperfilm MP (GE Healthcare) for 1 week.
For matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry, axonemal tubulin was purified from Tetrahymena cilia and C-terminal peptides were obtained as described previously (48). Peptide solutions were mixed 1:1 (vol/vol) with a saturated solution of 2,5-dihydroxybenzoic acid (Aldrich) in 0.1% aqueous trifluoroacetic acid. Mass spectra were acquired in the linear ion mode on a MALDI-TOF mass spectrometer (Voyager-STR; Perseptive Biosystems, Inc., Framingham, MA) under conditions described earlier (48).
RESULTS
Subtypes of microtubules in Tetrahymena differ in the maximal length of the polyglutamyl side chain.
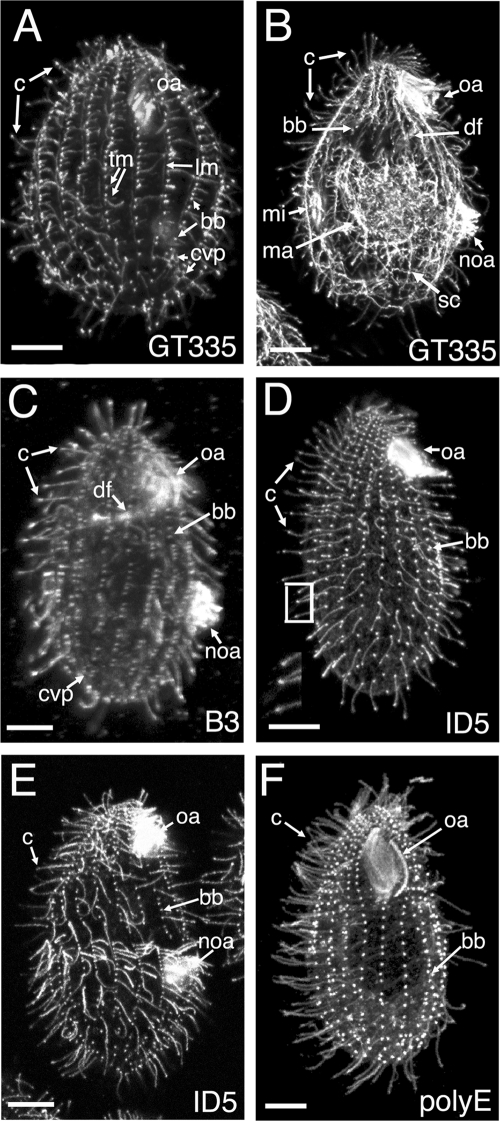
Throughout its life cycle, Tetrahymena assembles 18 types of microtubules, including those forming spindles, cilia and basal bodies, cortical arrays, and nuclear and cell body networks (19). Eleven locations of microtubules can be resolved in vegetatively growing cells by immunofluorescence (see Table S1 in the supplemental material); the remaining seven types of microtubules are found in conjugating cells and were not studied here. Microtubules of Tetrahymena are known to be glutamylated (6, 48). To test whether the polyglutamyl side chain length is regulated spatially, we stained vegetatively growing Tetrahymena cells with antibodies that recognize different glutamylated tubulin epitopes. The GT335 MAb recognizes a branch point of a glutamyl side chain consisting of at least one glutamate on either α- or β-tubulin (63) (see the footnote to Table S1 in the supplemental material). GT335 labeled all of the microtubule locations in vegetative cells (Fig. 1A and B). Thus, all of the microtubule locations in vegetative Tetrahymena contain at least monoglutamylated isoforms of either α- or β-tubulin. The B3 MAb recognizes glutamyl side chains in the context of the primary sequence of the CTT of either α- or β-tubulin (22, 31). Based on the sequences of the single conventional α-tubulin (Atu1p) and β-tubulin (Btu1/2p) and the B3 epitope mapping (31) in Tetrahymena, the B3 MAb is predicted to recognize side chains consisting of two or more glutamates (see the footnote to Table S1 in the supplemental material). B3 labeled 4 out of 11 locations, namely, cilia, basal bodies, contractile vacuole pores (CVP), and deep fiber microtubules (Fig. 1C). Thus, these microtubules likely contain biglutamyl or longer side chains. The ID5 MAb recognizes an XEE peptide ending with an α-carboxylate group (53). This epitope is present on glutamyl side chains of three or more glutamates (53). In many organisms, including vertebrates, the XEE epitope is also present at the C terminus of the primary chain of α-tubulin that has lost its terminal Y due to detyrosination and ends with GEE (60). However, in Tetrahymena, the only conventional α-tubulin, Atu1p, ends with Y and undergoes detyrosination in vivo (48) but the penultimate amino acid is G. Thus, in Tetrahymena, ID5 is predicted to recognize only glutamyl side chains of three or more glutamates. ID5 exclusively labeled cilia and basal bodies, indicating that only these two types of microtubules have tubulin side chains made of three or more glutamates (Fig. 1D and E). The polyE antibodies, originally generated against a Cys-E9 peptide (54), produced a pattern similar to that of ID5 (Fig. 1F) and thus most likely recognize side chains of at least three glutamates. Immunofluorescence data, taken together, led us to infer that microtubules that are labeled exclusively by GT335 (intracytoplasmic, nuclear, and cortical microtubules other than basal bodies) have only monoglutamylated tubulins. In two locations, CVP and the oral deep fiber, tubulin is detectable by B3 and GT335, but not polyE and ID5, antibodies and thus these tubulin isoforms likely have side chains limited to two glutamates. Cilia and basal bodies are the only structures that are labeled by ID5 and polyE and consequently are the only locations of tubulin with triglutamyl (or longer) side chains.
FIG. 1.
Distribution of glutamylated tubulin isoforms in Tetrahymena revealed by immunofluorescence with MAbs that recognize distinct glutamylated epitopes, i.e., GT335 (A and B), B3 (C), and ID5 (D and E), or with polyclonal polyE antibodies (F). (A and B) GT335 recognizes all types of microtubular systems, including intracytoplasmic, cortical, nuclear, and ciliary microtubules. The cell shown in panel B is approaching division; the new oral apparatus, micronuclear mitotic spindle, and intramacronuclear microtubules involved in amitosis are detected. (C) B3 labels basal bodies, cilia, oral deep fiber, and CVP. (D and E) Tubulin isoforms detected by ID5 are present only in basal bodies and cilia. In panel D, the inset shows a higher magnification of cilia in the boxed area. The cell shown in panel E is approaching cytokinesis. (F) PolyE antibodies label a set of structures identical to that labeled by the ID5 antibody (compare with panels D and E). Abbreviations: bb, basal body; c, cilia, cvp, CVP; df, deep fiber; lm, LMs; tm, transverse microtubules; ma, macronucleus; mi, micronucleus; noa, new oral apparatus; oa, oral apparatus; sc, subcortical microtubules. Bars = 10 μm.
Assuming that the localization data reported above are not affected by differential epitope masking, it appears that, in Tetrahymena, the maximal length of glutamyl side chains is regulated in a location-specific and possibly microtubule type-specific manner.
Ttll1p and Ttll9p glutamylases preferentially modify α-tubulin but differ in enzymatic properties and subcellular targeting.
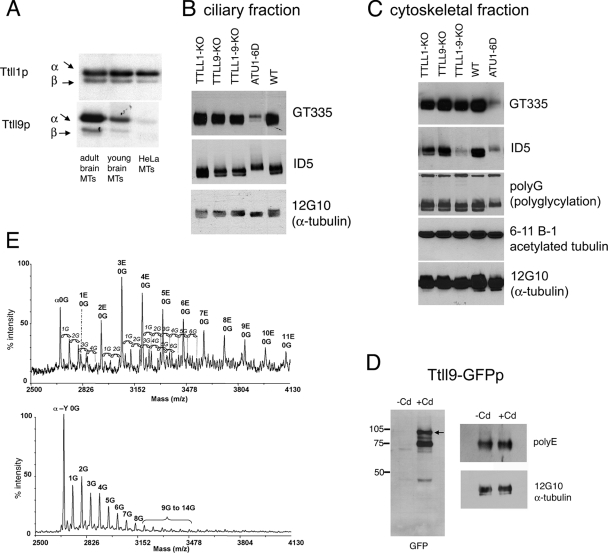
Tubulin glutamylases belong to the family of TTL (15) domain-carrying proteins named TTLL proteins (28). Mammals have 13 TTLLs (TTLL1 through TTLL13). Previous studies have identified murine TTLL4, TTLL5, TTLL6, TTLL7, TTLL11, and TTLL13 as distinct tubulin glutamylases that differ in the ability to either initiate or elongate side chains and prefer either α- or β-tubulin (27, 28, 57). The TTLL1 and TTLL9 proteins are evolutionarily related to the already well-characterized tubulin glutamylases (see Fig. S1 in the supplemental material). In fact, the discovery of glutamylating TTLLs was based on the observation that the murine TTLL1 copurified with α-tubulin-specific polyglutamylation activity during the fractionation of brain tissue (28). TTLL9 is closely related evolutionarily to TTLL1 (see Fig. S1 in the supplemental material). However, attempts to detect glutamylation activities associated with mammalian TTLL1 and TTLL9 expressed either in mammalian cells (57) or in Escherichia coli (J.V., K.R., and C.J., unpublished data) were not successful. Here, we characterize Ttll1p and Ttll9p, the Tetrahymena orthologs of mammalian TTLL1 and TTLL9 (see Fig. S1 in the supplemental material). We overproduced these two enzymes in Tetrahymena and purified enzyme-enriched fractions for in vitro glutamylation assays. The enzymatic mechanism of microtubule glutamylation involves two distinct steps: initiation and elongation (46, 50, 63). Some mammalian glutamylases are already known to have a mainly chain-initiating activity, while other enzymes primarily elongate side chains (57). To distinguish between initiating and elongating activities, we used three types of microtubule substrates which have high, medium, or low levels of preexisting glutamylation, respectively: microtubules made of (i) adult mouse brain tubulin, (ii) young mouse brain tubulin, and (iii) HeLa cell tubulin (5, 51). A GFP-Ttll1p-enriched fraction preferentially modified α-tubulin and used all three microtubule substrates about equally (Fig. 2A, top). The strong ability to modify HeLa microtubules indicates the presence of a major initiating activity. In vivo, GFP-Ttll1p localized primarily to the basal bodies of oral and somatic cilia (Fig. 3A to C), as well as to the oral deep fiber and CVP (28). Overproduction of Ttll1p in Tetrahymena did not change the pattern of glutamylation seen in immunofluorescence for either the ID5 MAb (Fig. 3B; compare to 1D and E) or the GT335 MAb (results not shown). Earlier, we found that overproduction of Ttll1p did not alter the total levels of tubulin glutamylation in vivo or in cell extracts, probably because this enzyme exists in a complex with another subunit(s) that is required for activity, as likely is also the case for the murine ortholog (28).
FIG. 2.
Deletion of TTLL1 and TTLL9 and mutations of polymodification sites on α-tubulin lead to loss of glutamylation in vivo. (A) Fluorographic analysis of products of in vitro glutamylation reactions with partially purified fractions of either GFP-Ttll1p (top) or GFP-Ttll9p (bottom). Different mammalian tubulin substrates were used as indicated. (B and C) Western blotting analysis of Tetrahymena cilia (B) and cytoskeletal fractions (C). The following antibodies were used: GT335 and ID5, anti-glutamylated tubulin MAbs; polyG, anti-polyglycine; 6-11-B1, anti-acetylated (K40) α-tubulin; 12G10, anti-α-tubulin MAbs. Note that under the gel conditions used, Tetrahymena α-tubulin migrates faster than β-tubulin, in contrast to the mammalian tubulin migration pattern shown in panel A. In addition, Tetrahymena tubulin subunits display a spread pattern of reactivity with the antiglutamylation and -glycylation antibodies which can be accounted for by the presence of series of isoforms with increasing degrees of modification (visible in the mass spectra in panel E), resulting in considerable apparent molecular mass heterogeneity. WT, wild type. (D) Western blot assay of cilium protein from GFP-Ttll9p cells (either induced or not induced with cadmium) analyzed with anti-GFP antibodies (the arrow indicates the expected position of the full-size protein). The values on the left are molecular sizes in kilodaltons. (E) Mass spectra of CTTs of axonemal α-tubulin isolated from cilia of Tetrahymena cells that are either wild type (top) or lack TTLL1 and TTLL9 (bottom) and were purified by arginine-Sepharose chromatography. Tetrahymena α-tubulin peptides corresponding to the CTT of the protein and starting at residue Asp424 are detected. The numbers of posttranslationally added glutamates (1E, 2E, etc.) and glycine residues (1G, 2G, and so on) and the detyrosinated form of α-tubulin (α-Y) are indicated.
FIG. 3.
Localization of GFP fusions of Ttll1p and Ttll9p. Double fluorescence confocal images of cells overexpressing either GFP-Ttll1p (A to C) or GFP-Ttll9p (D to I), showing a direct GFP signal (A, D, and G) or immunofluorescence signal with ID5 (B and E) or GT335 (H) anti-glutamylated tubulin antibodies. Panels C, F, and I are merged images. In Ttll9p-overexpressing cells (D to F), the small arrows point to cilia which have areas of increased glutamylation revealed by ID5. Note the colocalization of foci of increased glutamylation with the GFP-Ttll9p signal. In panels E and F, the insets show a higher magnification of cilia in the boxed area. Note that the cilia that accumulate GFP-Ttll9p have a “comet” of ID5 signal below the ciliary tip. Abbreviations: bb, basal body; noa, new oral apparatus; oa, oral apparatus. Bar = 10 μm.
Partially purified Ttll9p also preferentially modified α-tubulin. The activity was noticeably higher with microtubules made of adult brain tubulin compared to young brain tubulin and barely detectable with HeLa cell microtubules (Fig. 2A, lower panel). Thus, Ttll9p is associated mainly with a chain-elongating activity on α-tubulin. GFP-Ttll9p localized primarily to a subset of locomotory cilia, but a faint signal was also seen at oral and somatic basal bodies (Fig. 3D and G). Overproduction of GFP-Ttll9p enhanced the reactivity of ciliary microtubules to ID5 (Fig. 3D to F) but not to GT335 antibodies (Fig. 3G to I), in agreement with a chain-elongating activity. Areas of increased ID5 labeling were located just below the tips of cilia and colocalized with the sites of accumulation of GFP-Ttll9p (Fig. 3D to F; compare to 1D). Western blotting of cilia of cells overexpressing GFP-Ttll9p (Fig. 2D) showed a mild increase in the levels of polyE but not GT335 epitopes, again consistent with a chain-elongating activity of Ttll9p in vivo.
Loss of function of Ttll1p and Ttll9p affects the levels and patterns of tubulin glutamylation in vivo.
As shown above, Ttll1p and Ttll9p both preferentially modify α-tubulin. To shed light on the potential functional interactions between Ttll1p and Ttll9p, we disrupted these genes by DNA homologous recombination. We obtained viable and motile Tetrahymena cells lacking TTLL1, TTLL9, or both genes. Deletion of TTLL1 caused a strong reduction in the level of tubulin glutamylation in the basal bodies, based on immunofluorescence with the GT335 (results not shown) and ID5 antibodies (see Fig. S2 in the supplemental material). These results are consistent with the preferred localization of GFP-Ttll1p to the basal bodies (Fig. 3A). The TTLL9 knockout cells did not change the pattern of glutamylation seen in immunofluorescence (see Fig. S2 in the supplemental material). Based on Western blot assays, loss of TTLL1 alone reduced the levels of ID5 epitopes in the whole cytoskeletons (Fig. 2C) and loss of TTLL9 alone reduced the levels of ID5-reactive tubulins in cilia, primarily in the faster-migrating portion of the tubulin region that is enriched in α-tubulin (Fig. 2B). The double-knockout cells showed a loss of ID5 glutamylation epitopes that was greater than anticipated based on the sum of the two single-gene knockouts in cytoskeletons (Fig. 2C) but not in cilia (Fig. 2B). These data suggest that synergistic interactions occur between Ttll1p and Ttll9p in the cell body but not in cilia. The levels of other PTMs such as tubulin glycylation (47) or acetylation (37) appeared unaffected based on the analysis of the cytoskeletons (Fig. 2C).
Previously, we used MALDI-TOF mass spectrometry of total tubulin CTT peptide pools to characterize glutamylated forms of ciliary α-tubulin. The spectra obtained for wild-type axonemal tubulin showed that glutamylation occurs predominantly in α-tubulin and that it is a major modification, with up to 11 glutamates per CTT and a triglutamylated isoform as the most abundant species. In these peptide pools, glycylation was less extensive, reaching up to six G's per CTT, and the major fraction was nonglycylated (48) (Fig. 2E, top panel). Given that Ttll1p and Ttll9p modify predominantly α-tubulin in vitro, mass spectrometry of CTT pools was used to evaluate whether lack of these enzymes affects the levels of glutamylation on α-tubulin. The spectrum of double-knockout tubulin CTT peptides revealed exclusively glycylated isoforms of α-tubulin (up to 14 G's per peptide; Fig. 2E, bottom). These data are consistent with a substantial reduction in glutamylation on α-tubulin. However, in a Western blot assay of ciliary tubulins, a sizeable fraction of glutamylated epitopes was detected in the faster-migrating portion of the tubulin region, especially with the GT335 antibody (Fig. 2B, top panel). The absence of detection of some glutamylated peptides in the mass spectra is most probably due to the low abundance of the glutamylated forms and the difficulty in ionizing these highly acidic peptides in complex peptide mixtures, as previously observed (48). In conclusion, the mass spectrometry and Western blotting data, taken together, indicate that in the TTLL1 and TTLL9 double knockout, a decrease in the levels of glutamylation on α-tubulin has occurred.
To determine the role of polymodification sites on α-tubulin, we used site-directed in vivo mutagenesis (25) to construct a Tetrahymena strain, ATU1-6D, in which all six glutamates within the CTT of Atu1p (the only conventional α-tubulin in Tetrahymena) (38) were replaced with aspartates to preserve the charge but prevent polymodifications. The ATU1-6D cells are viable and motile, indicating that polymodifications on α-tubulin are not essential. In Western blot assays, there was a dramatic loss of both GT335 and ID5 epitopes in the cytoskeletons and cilia of ATU1-6D cells (Fig. 2B and C). The fast-migrating α-tubulin-enriched band completely disappeared, consistent with the anticipated absence of polymodifications acceptor sites in the CTT of α-tubulin in ATU1-6D cells. The ATU1-6D cells also showed a reduction in the levels of tubulin glycylation (Fig. 2C), which was expected since this PTM also occurs on glutamates of the CTT domain (59). Less expected was the fact that the signal of GT335 and ID5 also decreased in the upper portion of the tubulin doublet that is enriched in β-tubulin (Fig. 2C). Moreover, the electrophoretic mobility of the bulk of polyglutamylated forms of β-tubulin (ID5 positive) was slower in the ATU1-6D sample, possibly due to compensatory hyperelongation of glutamyl side chains or deposition of other, unknown, PTMs (Fig. 2B and C). This suggests that the mutations of modification sites on the CTT of α-tubulin also affect the levels of these modifications on β-tubulin, possibly by long-range effects across the microtubule lattice. We have already described such interactions between α- and β-tubulin for other CTT mutants (48).
Ttll1p and Ttll9p glutamylase enzymes and polymodification sites on α-tubulin are required for normal growth, phagocytosis, and sensitivity to antitubulin drugs.
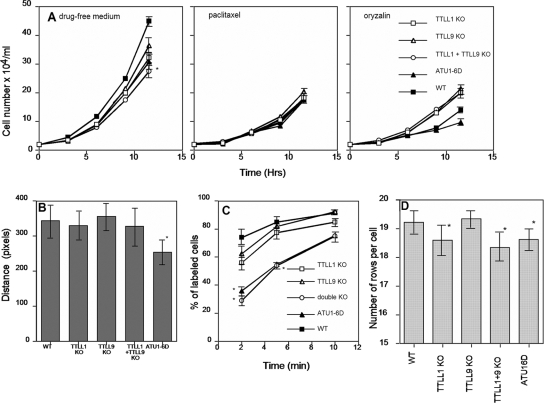
The single-knockout TTLL strains grew a little slower than the wild type, but the double-knockout cells showed a significant (P = 0.017) 20% increase in generation time (Fig. 4A). In the presence of 15 μM paclitaxel, the growth rates were reduced to similar levels in all of the strains tested (Fig. 4A). Thus, the advantageous function(s) that TTLL1 and TTLL9 provide may require normal dynamicity of microtubules. Surprisingly, the knockout strains grew slightly faster than wild-type cells in the presence of 10 μM oryzalin, a drug that depolymerizes microtubules and binds to α-tubulin (39). Thus, glutamylation on α-tubulin could directly or indirectly decrease the affinity of oryzalin for tubulin or protect microtubules from oryzalin-induced depolymerization.
FIG. 4.
(A) Growth curves of strains at 30°C in SPP medium or SPP supplemented with 15 μM paclitaxel or 10 μM oryzalin. Data represent mean values from three experiments and two experiments for oryzalin. For clarity, error bars are shown only for the last set of measurements. In panel A (left panel), the difference in the calculated generation time was significant with 95% confidence only between wild-type (WT) and double-knockout (KO) cells (P = 0.017). (B) Graph showing the rate of whole-cell motility expressed as the distance traveled by a cell within the time needed to collect five frames. Only the difference between wild-type and ATU1-6D cells was statistically significant (P < 0.0001). (C) Results of a phagocytosis assay. Averages from three experiments are shown, and 100 cells were scored for each time point. The percentage of cells with labeled (India ink) food vacuoles is indicated as a function of time after the addition of India ink. For the 2- and 5-min readouts, the differences between results obtained for wild-type and either double-knockout (P = 0.0037) or ATU1-6D (P = 0.005) cells were statistically significant. (D) Effects of gene knockouts on the average number of ciliary rows. Strains were analyzed at about 10 generations after their last mating. The results are mean values obtained for 50 cells. The differences between the results obtained with wild-type, TTLL1-null, and double-knockout or ATU1-6D cells were statistically significant (P = 0.001). All error bars represent standard deviations.
Tetrahymena cells use locomotory cilia for motility and oral cilia to direct food into a phagocytic vesicle. The rate of whole-cell motility was not significantly reduced in the knockout cells (Fig. 4B). However, the ATU1-6D cells showed a significant reduction in movement velocity, suggesting that either additional glutamylation (catalyzed by enzymes other than Ttll1p or Ttll9p) or glycylation on α-tubulin (or both) contributes to ciliary motility. Phagocytosis occurs inside the oral apparatus and requires motile oral cilia. While single-knockout cells showed only a mild reduction in the rate of phagocytosis (Fig. 4C), a profound defect was observed in double-knockout and ATU1-6D cells. After 5 min of exposure to India ink particles, about 80% of wild-type cells had labeled food vacuoles while only ∼43% of double-knockout cells did (P = 0.037). To conclude, the consequences of the loss of function of Ttll1p and Ttll9p indicate that both proteins work synergistically to regulate cell multiplication and phagocytosis.
Ttll1p or Ttll9p are required for normal assembly of cortical microtubules and maturation of basal bodies.
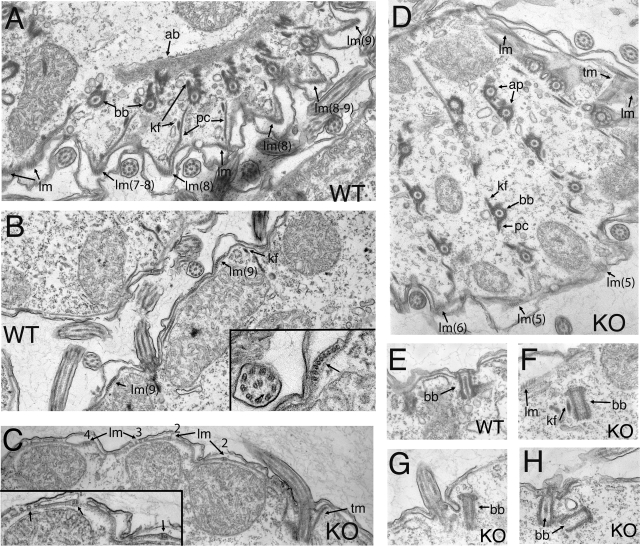
Since the phenotypic analyses described above showed synergistic interactions between Ttll1p and Ttll9p, we analyzed the ultrastructure of double-knockout cells. The cross sections of cilia, basal bodies, and postciliary and transverse microtubule bundles appeared normal (Fig. 5D). However, we observed an abnormal organization of longitudinal microtubule (LM) bundles (lm, Fig. 5C and D). LMs run parallel to basal body rows and are located between the epiplasm and alveolar sacs (44). The LM bundle is composed of partly overlapping microtubules arranged with an average of eight microtubules in the cross section (Fig. 5B, insert). The average number of microtubules in LMs in the TTLL1 and TTLL9 knockout cells was reduced by 34% (Fig. 5C, insert; the averages were 5.45 ± 1.73 [n = 47] for knockouts and 8.25 ± 2.1 [n = 28] for the wild type; P < 0.0001).
FIG. 5.
Transmission electron microscopy study of wild-type (A, B, and E) and TTLL1 and TTLL9 knockout cells (C, D, and F to H). (A) Section of the cortex at the apical part of a wild-type (WT) cell. (B) Section of the wild-type cortex. Note multiple cross sections of LMs and the organization of the microtubules in the bundle (arrow in inset). (C) Section of the double-knockout (KO) cell cortex showing fewer microtubules within the LM bundles (arrows; compare with panel B). (D) Section of apical cortex in the double-knockout cell showing a reduced number of microtubule profiles in LM bundles (compare with panel A). (E to H) Sections of basal bodies of wild-type (E) and TTLL1 and TTLL9 knockout cells (F to H). In panels F to H, note the presence of basal bodies that have an appearance of mature structures but are not anchored to the plasma membrane. In panels G and H, nascent basal bodies are present near template (ciliated) basal bodies. Abbreviations: ab, apical band; ap, apical pair of basal bodies; bb, basal body; kf, kinetodesmal fiber; lm, LMs; pc, postciliary microtubules; tm, transverse microtubules.
Although the cross sections of mature basal bodies appeared normal in the TTLL1 and TTLL9 knockouts, the location and morphology of some daughter basal bodies were abnormal (Fig. 5F to H). In wild-type cells, a new basal body forms anterior and at a right angle to the old basal body “template” (3). The nascent basal body elongates, rotates upward, and docks at the plasma membrane in front of the template basal body. The docked basal body undergoes maturation, including elongation and the formation of the axial granule at its top and accumulation of electron-dense material in the lumen (Fig. 5E). In the double-knockout cross sections, we found basal bodies that were fully elongated and closed by a terminal plate but were not docked at the plasma membrane, and their lumina had reduced density (Fig. 5F to H). Sections showing nascent basal bodies are rare, even in wild-type cells, and this prevented us from quantifying this phenotype, but to our knowledge, such mature but non-plasma membrane-anchored basal bodies have not been reported for wild-type cells. With the reduced efficiency of basal body maturation, the rows of basal bodies could shorten and some rows could even be lost (10, 11, 18). Within a growing wild-type population, a vast majority of cells maintains the number of ciliary rows within the so called “stability range” of 18 to 20 (17, 40). However, TTLL1-null and double-knockout cells had, on average, 1 row less per cell (Fig. 4D), with a sizeable fraction of cells with 17 rows (12% for the double knockout compared to 0% for the wild type [n = 50]). During an additional 150 generations of exponential growth, the number of rows below the normal range (<17) was 78% in double-knockout cells and 70% in TTLL1 knockout cells, compared to only 12% in the wild type (n = 50). The absence of a synergistic interaction between TTLL1 and TTLL9 indicates that Ttll1p alone contributes to basal body propagation, which is consistent with its localization.
DISCUSSION
Our studies reveal that in Tetrahymena, microtubule glutamylation is spatially regulated. The immunofluorescence data suggest that the maximal length of glutamyl side chains is regulated according to the microtubule organelle type, from monoglutamylation in most locations to biglutamylation or triglutamylation (and longer side chains) in selected organelles such as basal bodies and cilia. It appears that the observed differences in the patterns of glutamylation cannot be simply explained by differential longevity of microtubules. For example, in the cell cortex of Tetrahymena, only basal bodies and axonemes have triglutamylated or longer side chains, while all cortical microtubules, including the transverse or postciliary ribbons, are stable polymers that turnover slowly (55). A similar pattern of side chain length regulation was seen for the related polymeric PTM tubulin glycylation in the ciliate Paramecium, where intracytoplasmic microtubules have shorter side chains compared to cortical and axonemal microtubules (7). Data presented here indicate that preferential targeting of specific glutamylases to specific sites is a major factor that contributes to differential glutamylation. In particular, restricted targeting of glutamylases that have an elongating activity, such as Ttll6Ap on β-tubulin (57; unpublished observations) and Ttll9p on α-tubulin, could spatially regulate side chain length. As glutamylation is a reversible reaction in mammalian cells (2), it is likely that localized deglutamylation activity (which remains to be identified) is also involved.
By charge-conserving mutations of all six glutamate residues (to aspartates) in the CTT of the single conventional α-tubulin of Tetrahymena, ATU1p, we show that glutamylation, as well as glycylation, on α-tubulin is not essential. We showed earlier that the sites of polymodifications on the CTT of β-tubulin are essential in Tetrahymena (48, 55, 56, 64).
We have identified two conserved tubulin glutamylases, Ttll1p and Ttll9p, that preferentially modify α-tubulin. TTLL1- and TLLL9-type enzymes are highly conserved but are absent from higher plants and fungi (see Fig. S1 in the supplemental material). Thus, it is possible that these two enzymes have coevolved with cilia and centrioles/basal bodies. The localization of Ttll1p to basal bodies in vivo and our functional analyses implicate Ttll1p in the process of maturation and maintenance of duplicated basal bodies. Tagged Ttll9p localized primarily to cilia in Tetrahymena, and a TTLL9 protein was detected in the flagellar proteome of Chlamydomonas (42). We show that loss of function of both glutamylase enzymes leads to phenotypic consequences that are as severe as or less severe than those seen in the strain that lacks polymodifications sites on the CTT of α-tubulin. It is therefore unlikely that Ttll1p and Ttll9p glutamylate other important substrates in addition to Atu1p α-tubulin.
Our in vitro assays indicate that Ttll1p is a strong initiase, while Ttll9p is an elongase for α-tubulin. However, the results of some phenotypic assays (e.g., growth and phagocytosis) indicate that the two enzymes act synergistically rather than sequentially. To reconcile these data, we propose that Ttll9p elongates the side chains that are initiated at sites distinct from those initiated by Ttll1p.
We observed a synergistic effect of gene knockouts on the rates of cell multiplication and phagocytosis. Because the two proteins, Ttll1p and Ttll9p, have largely nonoverlapping localizations, it is not obvious how they can act synergistically. The reduced rate of multiplication could be mainly a consequence of slow phagocytosis, which is known to be required for optimal growth under conditions similar to those we have used (4). Ttll9p could contribute to the observed phenotypes mainly by affecting the beating rate or the waveform of oral cilia. In addition to basal bodies, Ttll1p localizes to the deep fiber, a bundle of microtubules that originates near the basal bodies of the oral apparatus (28). Microtubules forming deep fiber could participate in the motor-driven transport of food vacuoles from the oral cavity based on what has been proposed for similar microtubules in Paramecium (13). Alternatively, the deep fiber microtubules could mediate the delivery of membrane vesicles to regenerate the plasma membrane removed by phagocytosis. Thus, despite their nonoverlapping localizations, the two proteins could act synergistically by enhancing the efficiency of different steps in phagocytosis.
Not all of the phenotypic effects of gene knockouts corresponded to the detectable localizations of tagged Ttll1p or Ttll9p. Specifically, the effect of gene knockouts on the number of microtubules in LM bundles was unexpected since neither tagged Ttll1p nor Ttll9p was detectably associated with LMs. LMs do contain monoglutamylated tubulin (Fig. 1A; see Table S1 in the supplemental material). It is possible that either Ttll1p or Ttll9p associates with LMs but is present at a level below the limit of detection by immunofluorescence. Alternatively, the effect of lack of glutamylation on LMs could be an indirect consequence of the lack of proper function of basal bodies that are aligned on one side of the LMs. It is remarkable, however, that in the knockout strains, only LM bundles and not other types of cortical microtubules, such as PCs and TMs, were affected.
We show here that Ttll1p and Ttll9p are required for the proper biogenesis of specific microtubular organelles. Mutants lacking both Ttll1p and Ttll9p have fewer microtubules in LMs and occasionally fail to dock basal bodies to the plasma membrane. Campbell and colleagues found that a mutation in murine PGs1, a noncatalytic subunit that associates with TTLL1 (52), leads to either lack of assembly or gross abnormalities in the assembled sperm axonemes (8). An RNA interference-based knockdown of TTLL7 β-tubulin glutamylase inhibited the outgrowth of neurites in PC12 cells (27). A knockdown of TTLL6 β-tubulin inhibited the assembly of a subset of cilia in zebra fish (41). All of these data, together with our observations, indicate that glutamylation on α- and β-tubulin plays a conserved role as a major determinant of the process of assembly of complex microtubular systems such as microtubule bundles, axonemes, and centrioles/basal bodies.
To conclude, we present here a functional characterization of two enzymes, TTLL1 and TTLL9, and show that both mediate glutamylation on α-tubulin. Taking our results together with the already published data, we conclude that there are at least five evolutionarily conserved types of tubulin glutamylases: TTLL1, TTLL4, TTLL5, TTLL6, and TTLL9. Including all paralogs, Tetrahymena has at least 13 predicted glutamylase proteins (see Fig. S1 in the supplemental material). We present evidence supporting a model implying that specific properties of microtubules are generated by multiple spatially restricted glutamylating enzymes that operate within the same cell, by showing that (i) the length of polyglutamyl side chains is microtubule type dependent, (ii) tubulin glutamylases have restricted subcellular locations, and (iii) deficiencies in enzymes affect a subset of microtubule types and microtubule-dependent functions.
It is not know yet how microtubule glutamylation changes the structural properties of microtubules. In the future, it will be important to determine whether glutamylation changes the intrinsic properties of microtubules (such as the polymer dynamics) or acts indirectly by regulating the affinity of microtubule-associated proteins and motors. The characterization of TTLL1 and TTLL9 as α-tubulin-specific enzymes and the creation of loss-of-function mutants as described here should be useful in future studies on the structural consequences of tubulin glutamylation on microtubules by both in vitro and in vivo approaches.
Supplementary Material
Acknowledgments
This work was supported by NSF grant MBC-033965 (to J.G.), NIH grant GM-26973 (to M.A.G.), Association de la Recherche contre le Cancer awards CR504/7817 and 3140 (to C.J.), French National Research Agency (ANR) award JC05_42022 (to C.J.), La Ligue contre le Cancer awards (to C.J. and K.R.), the statute grant to the Nencki Institute from the Polish Ministry of Science and Higher Education (to M. J.-D.), support from INSERM (to V.R.), and funds from the CNRS and the Université Paris-Sud (to M.-H.B. and N.L.).
We thank Henryk Bilski and Kazimierz Krawczyk of the Laboratory for Electron Microscopy at the Nencki Institute for excellent technical assistance, Iza Strzyzewska-Jowko for help with transmission electron microscopy fixation, and Pierre Le Maréchal of the IBBMC (CNRS, Orsay, France) for access to the MALDI-TOF mass spectrometer. We thank Joseph Frankel (University of Iowa) for insightful discussions. We thank Klaus Weber (Max Planck Institute, Goettingen, Germany) for ID5, Joseph Frankel for the 12G10 antibody (available from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa), and Donna Cassidy-Hanley (Cornell University) for the CU522 strain.
Footnotes
Published ahead of print on 27 June 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Allen, S. L. 1967. Genomic exclusion: a rapid means for inducing homozygous diploid lines in Tetrahymena pyriformis, syngen 1. Science 155575-577. [DOI] [PubMed] [Google Scholar]
- 2.Audebert, S., E. Desbruyères, C. Gruszczynski, A. Koulakoff, F. Gros, P. Denoulet, and B. Eddé. 1993. Reversible polyglutamylation of alpha- and beta-tubulin and microtubule dynamics in mouse brain neurons. Mol. Biol. Cell 4615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aufderheide, K. J., J. Frankel, and N. E. Williams. 1980. Formation and positioning of surface-related structures in protozoa. Microbiol. Rev. 44252-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basmussen, L., and E. Orias. 1975. Tetrahymena: growth without phagocytosis. Science 190464-465. [DOI] [PubMed] [Google Scholar]
- 5.Bobinnec, Y., M. Moudjou, J. P. Fouquet, E. Desbruyères, B. Eddé, and M. Bornens. 1998. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskelet. 39223-232. [DOI] [PubMed] [Google Scholar]
- 6.Bré, M. H., B. de Néchaud, A. Wolff, and A. Fleury. 1994. Glutamylated tubulin probed in ciliates with the monoclonal antibody GT335. Cell Motil. Cytoskelet. 27337-349. [DOI] [PubMed] [Google Scholar]
- 7.Bré, M. H., V. Redeker, J. Vinh, J. Rossier, and N. Levilliers. 1998. Tubulin polyglycylation: differential posttranslational modification of dynamic cytoplasmic and stable axonemal microtubules in Paramecium. Mol. Biol. Cell 92655-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, P. K., K. G. Waymire, R. L. Heier, C. Sharer, D. E. Day, H. Reimann, J. M. Jaje, G. A. Friedrich, M. Burmeister, T. J. Bartness, L. D. Russell, L. J. Young, M. Zimmer, D. E. Jenne, and G. R. MacGregor. 2002. Mutation of a novel gene results in abnormal development of spermatid flagella, loss of intermale aggression and reduced body fat in mice. Genetics 162307-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassidy-Hanley, D., J. Bowen, J. Lee, E. S. Cole, L. A. VerPlank, J. Gaertig, M. A. Gorovsky, and P. J. Bruns. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen-Shan, L. 1969. Cortical morphogenesis in Paramecium aurelia following amputation of the posterior region. J. Exp. Zool. 170205-227. [Google Scholar]
- 11.Chen-Shan, L. 1970. Cortical morphogenesis in Paramecium aurelia following amputation of the anterior region. J. Exp. Zool. 174463-478. [Google Scholar]
- 12.Chilcoat, N. D., S. M. Melia, A. P. Haddad, and A. P. Turkewitz. 1996. Granule lattice protein-1 (GRLP-1), an acidic, calcium-binding protein in Tetrahymena thermophila dense-core secretory granules, influences granule size, shape, content organization, and release but not protein sorting or condensation. J. Cell Biol. 1351775-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, J., N. Garreau de Loubresse, and J. Beisson. 1984. Actin microfilaments in Paramecium: localization and role in intracellular movements. Cell Motil. 4443-468. [DOI] [PubMed] [Google Scholar]
- 14.Eddé, B., J. Rossier, J.-P. Le Caer, E. Desbruyères, F. Gros, and P. Denoulet. 1990. Posttranslational glutamylation of α-tubulin. Science 24783-85. [DOI] [PubMed] [Google Scholar]
- 15.Ersfeld, K., J. Wehland, U. Plessmann, H. Dodemont, V. Gerke, and K. Weber. 1993. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 120725-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46101-111. [DOI] [PubMed] [Google Scholar]
- 17.Frankel, J. 1980. Propagation of cortical differences in Tetrahymena. Genetics 94607-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel, J., and N. E. Williams. 1973. Cortical development in Tetrahymena, p. 375-409. In A. M. Elliott (ed.), The biology of Tetrahymena. Dowden, Hutchinson, and Ross, Stroudsburg, PA.
- 19.Gaertig, J. 2000. Molecular Mechanisms of microtubular organelle assembly in Tetrahymena. J. Eukaryot. Microbiol. 47185-190. [DOI] [PubMed] [Google Scholar]
- 20.Gaertig, J., M. A. Cruz, J. Bowen, L. Gu, D. G. Pennock, and M. A. Gorovsky. 1995. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J. Cell Biol. 1291301-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaertig, J., Y. Gao, T. Tishgarten, T. G. Clark, and H. W. Dickerson. 1999. Surface display of a parasite antigen in the ciliate Tetrahymena thermophila. Nat. Biotechnol. 17462-465. [DOI] [PubMed] [Google Scholar]
- 22.Gagnon, C., D. White, J. Cosson, P. Huitorel, B. Eddé, E. Desbruyères, L. Paturle-Lafanèchere, L. Multigner, D. Job, and C. Cibert. 1996. The polyglutamylated lateral chain of alpha-tubulin plays a key role in flagellar motility. J. Cell Sci. 1091545-1553. [DOI] [PubMed] [Google Scholar]
- 23.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12543-548. [DOI] [PubMed] [Google Scholar]
- 24.Gorovsky, M. A., M.-C. Yao, J. B. Keevert, and G. L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. IX311-327. [DOI] [PubMed] [Google Scholar]
- 25.Hai, B., J. Gaertig, and M. A. Gorovsky. 2000. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62513-531. [DOI] [PubMed] [Google Scholar]
- 26.Hai, B., and M. A. Gorovsky. 1997. Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 941310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikegami, K., M. Mukai, J. Tsuchida, R. L. Heier, G. R. Macgregor, and M. Setou. 2006. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J. Biol. Chem. 28130707-30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janke, C., K. Rogowski, D. Wloga, C. Regnard, A. V. Kajava, J.-M. Strub, N. Temurak, J. van Dijk, D. Boucher, A. van Dorsselaer, S. Suryavanshi, J. Gaertig, and B. Eddé. 2005. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 3081758-1762. [DOI] [PubMed] [Google Scholar]
- 29.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23403-405. [DOI] [PubMed] [Google Scholar]
- 30.Jerka-Dziadosz, M., I. Strzyewska-Jowko, U. Wojsa-Lugowska, W. Krawczynska, and A. Krzywicka. 2001. The dynamics of filamentous structures in the apical band, oral crescent, fission line and the postoral meridional filament in Tetrahymena thermophila revealed by the monoclonal antibody 12G9. Protist 15253-67. [DOI] [PubMed] [Google Scholar]
- 31.Kann, M. L., S. Soues, N. Levilliers, and J. P. Fouquet. 2003. Glutamylated tubulin: diversity of expression and distribution of isoforms. Cell Motil. Cytoskelet. 5514-25. [DOI] [PubMed] [Google Scholar]
- 32.Kreitzer, G., G. Liao, and G. G. Gundersen. 1999. Detyrosination of tubulin regulates the interaction of intermediate filaments with microtubules in vivo via a kinesin-dependent mechanism. Mol. Biol. Cell 101105-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larcher, J. C., D. Boucher, S. Lazereg, F. Gros, and P. Denoulet. 1996. Interaction of kinesin motor domains with alpha- and beta-tubulin subunits at a tau-independent binding site. Regulation by polyglutamylation. J. Biol. Chem. 27122117-22124. [DOI] [PubMed] [Google Scholar]
- 35.Lechtreck, K.-F., and S. Geimer. 2000. Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil. Cytoskelet. 47219-235. [DOI] [PubMed] [Google Scholar]
- 36.Lefebvre, P. A. 1995. Flagellar amputation and regeneration in Chlamydomonas. Methods Cell Biol. 473-7. [DOI] [PubMed] [Google Scholar]
- 37.L'Hernault, S. W., and J. L. Rosenbaum. 1983. Chlamydomonas α-tubulin is posttranslationally modified in the flagella during flagellar assembly. J. Cell Biol. 97258-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGrath, K. E., S. M. Yu, D. P. Heruth, A. A. Kelly, and M. A. Gorovsky. 1994. Regulation and evolution of the single alpha-tubulin gene of the ciliate Tetrahymena thermophila. Cell Motil. Cytoskelet. 27272-283. [DOI] [PubMed] [Google Scholar]
- 39.Morrissette, N. S., A. Mitra, D. Sept, and L. D. Sibley. 2004. Dinitroanilines bind alpha-tubulin to disrupt microtubules. Mol. Biol. Cell 151960-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nanney, D. L. 1966. Corticotype transmission in Tetrahymena. Genetics 54955-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pathak, N., T. Obara, S. Mangos, Y. Liu, and I. A. Drummond. 2007. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol. Biol. Cell 184353-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pazour, G. J., N. Agrin, J. Leszyk, and G. B. Witman. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peris, L., M. Thery, J. Faure, Y. Saoudi, L. Lafanechere, J. K. Chilton, P. Gordon-Weeks, N. Galjart, M. Bornens, L. Wordeman, J. Wehland, A. Andrieux, and D. Job. 2006. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J. Cell Biol. 174839-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitelka, D. R. 1961. Fine structure and silver line and fibrillar systems of three tetrahyminid species. J. Protozool. 875-89. [Google Scholar]
- 45.Plessmann, U., I. Reiter-Owona, and K. F. Lechtreck. 2004. Posttranslational modifications of α-tubulin of Toxoplasma gondii. Parasitol. Res. 94386-389. [DOI] [PubMed] [Google Scholar]
- 46.Redeker, V., J.-P. Le Caer, J. Rossier, and J. C. Promé. 1991. Structure of the polyglutamyl side chain posttranslationally added to α-tubulin. J. Biol. Chem. 26623461-23466. [PubMed] [Google Scholar]
- 47.Redeker, V., N. Levilliers, J.-M. Schmitter, J.-P. Le Caer, J. Rossier, A. Adoutte, and M.-H. Bré. 1994. Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science 2661688-1691. [DOI] [PubMed] [Google Scholar]
- 48.Redeker, V., N. Levilliers, E. Vinolo, J. Rossier, D. Jaillard, D. Burnette, J. Gaertig, and M.-H. Bré. 2005. Mutations of tubulin glycylation sites reveal cross-talk between the C termini of α- and β-tubulin and affect the ciliary matrix in Tetrahymena. J. Biol. Chem. 280596-606. [DOI] [PubMed] [Google Scholar]
- 49.Reed, N. A., D. Cai, L. Blasius, G. T. Jih, E. Meyhofer, J. Gaertig, and K. J. Verhey. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 162166-2172. [DOI] [PubMed] [Google Scholar]
- 50.Regnard, C., S. Audebert, É. Desbruyères, P. Denoulet, and B. Eddé. 1998. Tubulin polyglutamylase: partial purification and enzymatic properties. Biochemistry 378395-8404. [DOI] [PubMed] [Google Scholar]
- 51.Regnard, C., É. Desbruyères, P. Denoulet, and B. Eddé. 1999. Tubulin polyglutamylase: isozymic variants and regulation during the cell cycle in HeLa cells. J. Cell Sci. 1124281-4289. [DOI] [PubMed] [Google Scholar]
- 52.Regnard, C., D. Fesquet, C. Janke, D. Boucher, É. Desbruyères, A. Koulakoff, C. Insina, P. Travo, and B. Eddé. 2003. Characterization of PGs1, a subunit of a protein complex co-purifying with tubulin polyglutamylase. J. Cell Sci. 1164181-4190. [DOI] [PubMed] [Google Scholar]
- 53.Rüdiger, A. H., M. Rüdiger, J. Wehland, and K. Weber. 1999. Monoclonal antibody ID5: epitope characterization and minimal requirements for the recognition of polyglutamylated alpha- and beta-tubulin. Eur. J. Cell Biol. 7815-20. [DOI] [PubMed] [Google Scholar]
- 54.Shang, Y., B. Li, and M. A. Gorovsky. 2002. Tetrahymena thermophila contains a conventional γ-tubulin that is differentially required for the maintenance of different microtubule organizing centers. J. Cell Biol. 1581195-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thazhath, R., M. Jerka-Dziadosz, J. Duan, D. Wloga, M. A. Gorovsky, J. Frankel, and J. Gaertig. 2004. Cell context-specific effects of the β-tubulin glycylation domain on assembly and size of microtubular organelles. Mol. Biol. Cell 154136-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thazhath, R., C. Liu, and J. Gaertig. 2002. Polyglycylation domain of β-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat. Cell Biol. 4256-259. [DOI] [PubMed] [Google Scholar]
- 57.van Dijk, J., K. Rogowski, J. Miro, B. Lacroix, B. Eddé, and C. Janke. 2007. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell 26437-448. [DOI] [PubMed] [Google Scholar]
- 58.Verhey, K. J., and J. Gaertig. 2007. The tubulin code. Cell Cycle 62152-2160. [DOI] [PubMed] [Google Scholar]
- 59.Vinh, J., J. I. Langridge, M.-H. Bré, N. Levilliers, V. Redeker, D. Loyaux, and J. Rossier. 1999. Structural characterization by tandem spectroscopy of the posttranslational modifications of tubulin. Biochemistry 383133-3139. [DOI] [PubMed] [Google Scholar]
- 60.Wehland, J., and K. Weber. 1987. Turnover of the carboxy-terminal tyrosine of α-tubulin and means of reaching elevated levels of detyrosination in living cells. J. Cell Sci. 88(Pt. 2)185-203. [DOI] [PubMed] [Google Scholar]
- 61.Westermann, S., and K. Weber. 2003. Post-translational modifications regulate microtubule function. Nat. Rev. Mol. Cell Biol. 4938-947. [DOI] [PubMed] [Google Scholar]
- 62.Wloga, D., A. Camba, K. Rogowski, G. Manning, M. Jerka-Dziadosz, and J. Gaertig. 2006. Members of the Nima-related kinase family promote disassembly of cilia by multiple mechanisms. Mol. Biol. Cell 172799-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolff, A., B. De Néchaud, D. Chillet, H. Mazarguil, É. Desbruyères, S. Audebert, B. Eddé, F. Gros, and P. Denoulet. 1992. Distribution of glutamylated α- and β-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59425-432. [PubMed] [Google Scholar]
- 64.Xia, L., B. Hai, Y. Gao, D. Burnette, R. Thazhath, J. Duan, M.-H. Bré, N. Levilliers, M. A. Gorovsky, and J. Gaertig. 2000. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 1491097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.