Figure 1. Scheme of the procedure for the selection of anti-immune complex phage and set up of Phage Anti-immune complex Assay (PHAIA).
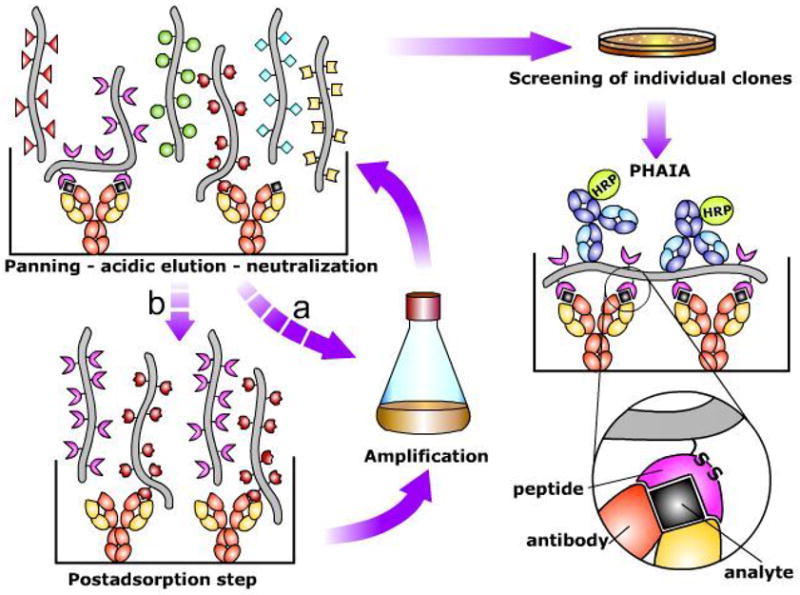
An anti-analyte antibody adsorbed onto the solid phase of an ELISA plate is used to capture the analyte (black square). A phage display peptide library is then added, and after several washing steps, specific phages recognizing the immune complex are eluted by addition of low pH buffer. The supernatant is neutralized, and the selected subpopulation of phages can be used directly for E. coli infection and phage amplification (a), or submitted to an optional post adsorption step to eliminate phages with strong affinity for the uncombined antibody (b). After several rounds of selection (panning) individual clones are tested for differential binding to the immune complex and the free antibody by phage ELISA. Phage clones showing low reactivity with the empty antibody are selected for development of PHAIA. In this assay, the immune complex formation is detected by its reaction with the phage borne peptide, followed by an anti-pVIII phage coat protein antibody coupled to horse radish peroxidase (HRP). A possible scheme of the antibody-analyte-peptide trivalent interaction that allows development of PHAIA is shown in the lower right part of the figure. Note that the overall affinity of the peptide for the immune complex is postulated to be the result of their interaction with the analyte and the antibody. The relative role of these contributions is further discussed in the text.
