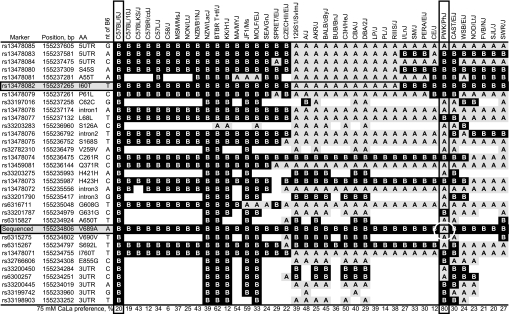
Fig. 5.
Tas1r3 single nucleotide polymorphism (SNP) analysis of 40 inbred strains of mice. Left columns give each marker's name, position (MGI, Build 37), and effect on amino acid (AA) sequence, if any. Column labeled “nt of B6” shows nucleotide present in C57BL/6J strain. In body of table, B = same nucleotide as B6; A = alternative to B6 (i.e., other nucleotide); blank = no genotype available. Strains are arranged from left to right so as to display similarities. PWK/PhJ and PWD/Ph (not shown) have the same haplotype; 129S1/SvImJ and 129P3/J (not shown) have the same haplotype. The I60T substitution (at 155,237,265 bp; shaded) is believed to be primarily responsible for determining responsiveness of Tas1r3 to saccharin. The critical SNP at “Sequenced” (shaded) is unique to the PWK/PhJ and PWD/PhJ strains (highlighted with circle) and may thus be responsible for these strains high calcium preferences. Preference data for 75 mM CaLa in bottom row are extracted from Ref. 43.