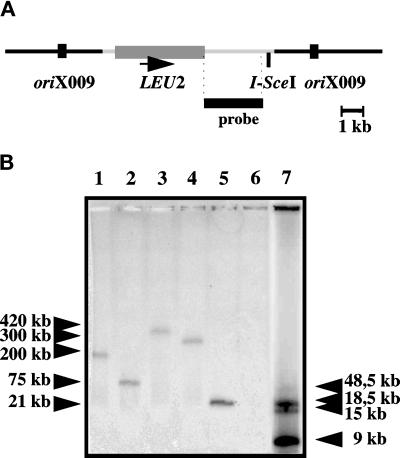
Figure 2.
Localization of the repeated oriX009 sequence in the genome of Y. lipolytica. (A) Map of a chromosomal integration at an oriX009 locus of the pINA989 containing oriX009 within its genomic environment, the LEU2 gene, and an I-SceI site. pBR322 vector sequences are shown in light gray. (B) Chromosomes from various transformants (lanes 1–5) were embedded in agarose plugs, digested with I-SceI, and separated under the following field inversion gel electrophoresis conditions: 1% pulsed field agarose (Bio-Rad, Bio-Rad, Hercules, CA) in 0.5× TAE buffer, run at 6 volts/cm for 14 h at 14°C with an angle of 106° and a variation of pulse from 0.41 sec to 2 min 6.52 sec. The DNA was transferred to a membrane and hybridized with a pBR322 probe (black box). A nontransformed strain is shown as a control in lane 6. The Raoul marker (Appligene, Heidelberg, Germany) is in lane 7. S. cerevisiae chromosomes and λ concatemers from Bio-Rad (not visible on the autoradiogram) were also used as molecular weight markers. As a comparison, the smallest chromosome among the six from Yarrowia is 2.6 Mb, and the largest is 4.9 Mb.