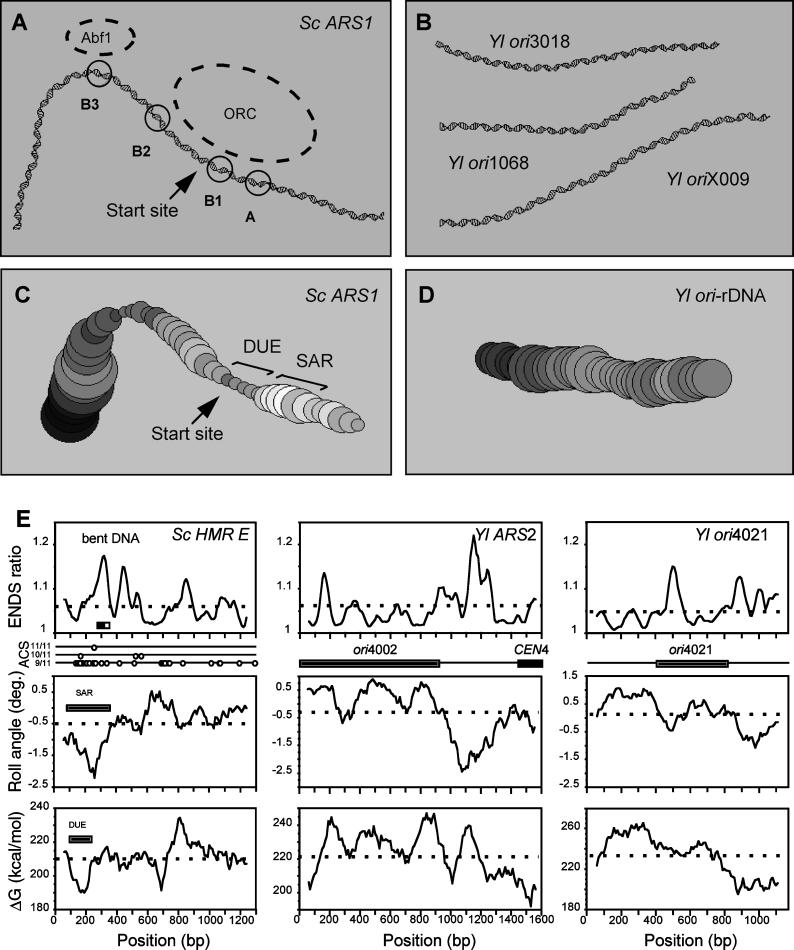
Figure 6.
Y. lipolytica origins do not contain structural motifs that are frequent in S. cerevisiae ARS elements. (A) S. cerevisiae ARS1 contains a strong, bent DNA region. The 2D projection of the 3D path of the molecule was calculated as described in MATERIALS AND METHODS. The position of the three elements A, B1, B2, and B3 (Marahrens and Stillman, 1992), the binding sites of ORC and ABF1 (Lee and Bell, 1997), and the site of initiation of DNA synthesis (Bielinsky and Gerbi, 1998) are indicated. (B) Y. lipolytica origins are not intrinsically curved. The 3D path of the minimal origin region of ori3018 (144 bp), ori1068 (125 bp), and oriX009 (159 bp) was calculated as described for ARS1. (C)Variation of helical stability and minor groove width along the S. cerevisiae ARS1 DNA fragment (350 bp). The free energy of the duplex or ΔG (207–227 kcal/mol; circles of increasing size) and the mean Roll angle (−1 to −0.1°; gray levels of increasing intensity) were calculated as described in MATERIALS AND METHODS for a 120-bp window sliding along the molecule. The position of the SAR mapped by Amati et al., 1988 (low Roll angle) and of the DUE (low ΔG) are indicated. (D) Spatial path and helical parameters of Y. lipolytica minimal ori-rDNA fragment (284 bp). The mean Roll angle varies from −0.3 to 0.5°, and the ΔG ranges from 215 to 226 kcal/mol. (E) Structural maps of large fragments containing S. cerevisiae HMRE ARS, Y. lipolytica ARS2, and Y. lipolytica ori4021. Like ARS1, the ACS of the HMRE ARS is flanked by a bent motif bearing binding sites for the auxiliary factors RAP1 (dark box) and ABF1 (open box) and by a DUE. The SAR fragment (Amati et al., 1988) maps perfectly to the region of low Roll angle. In all the graphs, the dotted lines correspond to the average value. Ori4021 is shown in its pBR322 environment, and the other two origins are at their chromosomal locus.