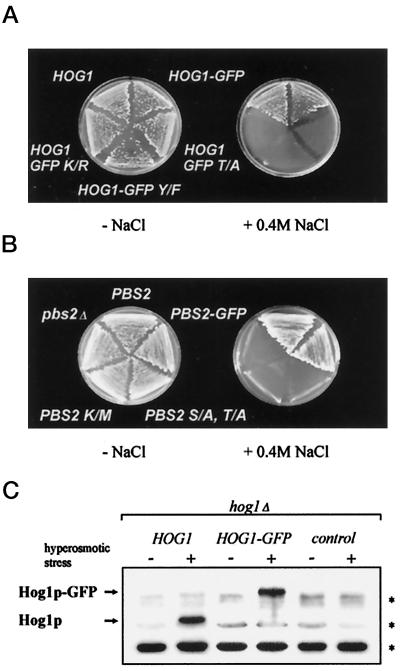
Figure 1.
Expression of Hog1–GFP and Pbs2–GFP complement hyperosmotic sensitivity of corresponding genomic mutants. (A) Strain K4327 (hog1Δ) was transformed with centromer plasmid pVR50 (HOG1), pVR65–WT (HOG1–GFP), pVR65–T/A (HOG1–GFP T/A), pVR65–Y/F (HOG1–GFP Y/F), or pVR65–K/R (HOG1–GFP K/R). The expression of HOG1 alleles was driven by their endogenous promoter. Transformants were grown on selective medium at 30°C without or with 0.4 M NaCl. (B) Strain VRY 10 (pbs2Δ) was transformed with pVR15 (PBS2), pVR15–GFP (PBS2–GFP), pVR20 (PBS2 S/A, T/A), or pVR15K/M (PBS2 K/M) and YCp22 (control plasmid, pbs2Δ). Expression of PBS2 alleles was driven by their endogenous promoter. Transformants were grown on selective medium at 30°C in the absence or presence of 0.4 M NaCl. (C) Antibody against an active human p38 MAPK specifically recognizes an activated form of yeast Hog1. The protein extracts from strain K4327 (hog1Δ) transformed with a centromer plasmid bearing a WT HOG1 (pVR50), HOG1–GFP fusion gene (pVR65–WT), or control plasmid (YCp111) were tested. Cells were grown in selective medium. For hyperosmotic stress, NaCl was added to a final concentration of 0.4 M for 5 min before preparation of protein extracts, which were analyzed by Western blotting using antibody against active p38 MAPK. Asterisks indicate the positions of a set of proteins from yeast crude extract (e.g., those with slightly higher mobility than Hog1) that are nonspecifically recognized by anti-active human p38 antibody.