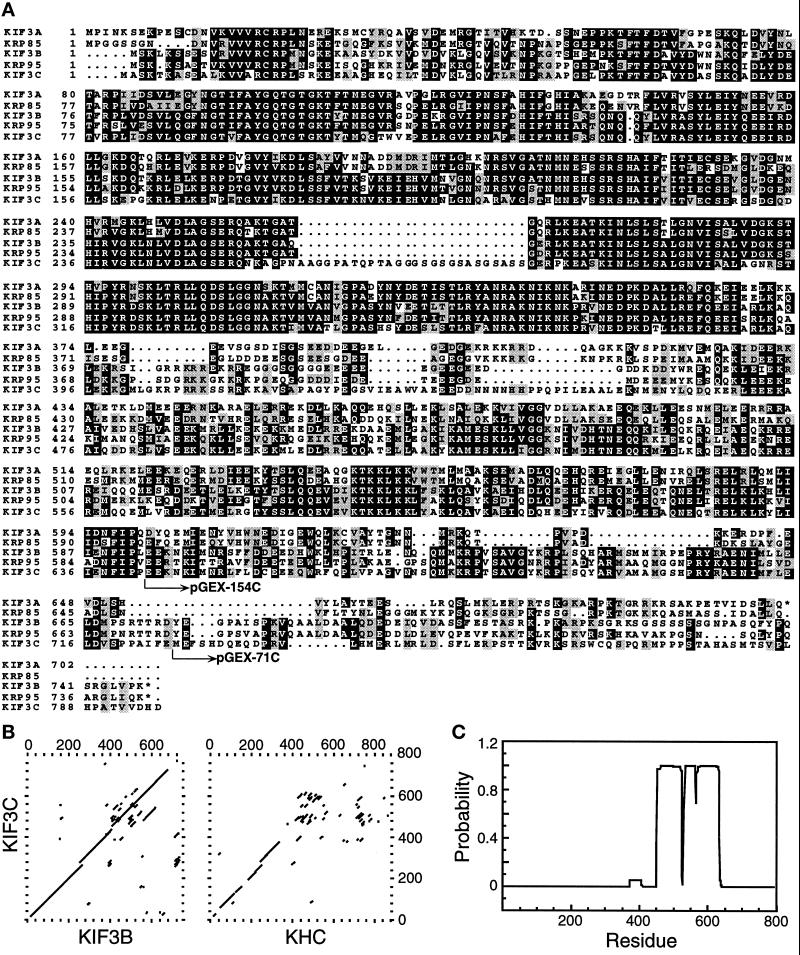
Figure 1.
Sequence analysis of KIF3C. (A) Predicted amino acid sequence alignment of KIF3C with KIF3A, KIF3B, KRP85, and KRP95. Identical amino acids, and conserved, but not identical amino acids, in the five polypeptides are shown in black and light boxes, respectively. pGEX-154C and pGEX-71C indicate the fusion points of GST protein and KIF3C protein regions, which were used for generating antigens and antibodies. The alignment was performed with the UWGCG software package (Devereux et al., 1984) and boxed by the program BOXSHADE 3.21 (http://ulrec3.unil.ch:80/software). These sequence data are available from EMBL/GenBank under accession number AF013116. (B) Dot matrix sequence comparisons of KIF3C with KIF3B and mouse KHC. Comparisons were done with the UWGCG program COMPARE (window = 30, stringency = 16), and were plotted with the program DOTPLOT. (C) The probability that each residue of KIF3C will participate in an α-helical coiled-coil structure is represented in a bar graph, as calculated by the algorithm of Lupas (Lupas et al., 1991).