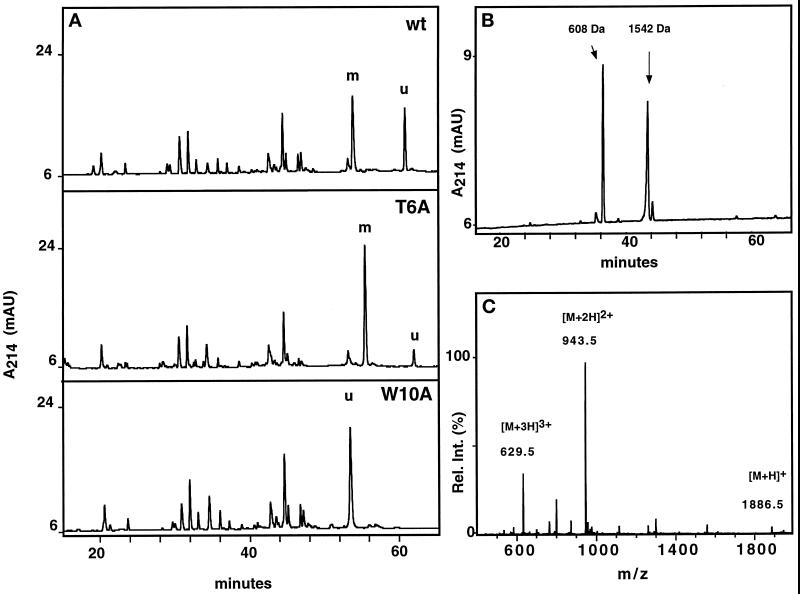
Figure 4.
Characterization of RNase 2.4 and its mutants. (A) Five micrograms of reduced and carboxymethylated RNase were digested at glutamic acid residues with endoproteinase Glu-C and fractionated by C8 reversed-phase LC-ESIMS. A 1-mm diameter column equilibrated in 95% solvent A (2% CH3CN, 0.05% TFA) and 5% solvent B (80% CH3CN, 0.045% TFA) was used. Peptides were eluted with a linear gradient of 5–40% solvent B at a flow rate of 50 μl/min. C-mannosylated (“m”) and unmodified (“u”) fragment −4 to 12 were assigned based on their molecular masses. The results obtained with wild-type RNase 2.4 (upper panel), mutant T6A (middle panel), and W10A (lower panel) are shown as representative examples. (B) Peptide −4 to 12 was digested with elastase and fractionated by reversed-phase LC-ESIMS. The peptide map obtained from the T6A mutant is shown as a representative example. Peptide 9–12 with unmodified Trp-10, 608 Da; peptide −4 to 8 with C-mannosylated Trp-7, 1542 Da. (C) ESIMS of the fragment −4 to 12 from RNase 2.4, W10A. The molecular mass of 1885 Da corresponds to that of the peptide with unmodified Trp.