Abstract
It has been many years since position-specific residue preference around the ends of a helix was revealed. However, all the existing secondary structure prediction methods did not exploit this preference feature, resulting in low accuracy in predicting the ends of secondary structures. In this study, we collected a relatively large data set consisting of 1860 high-resolution, non-homology proteins from the PDB, and further analyzed the residue distributions around the ends of regular secondary structures. It was found that there exist position-specific residue preferences (PSRP) around the ends of not only helices but also strands. Based on the unique features, we proposed a novel strategy and developed a tool named E-SSpred that treats the secondary structure as a whole and builds models to predict entire secondary structure segments directly by integrating relevant features. In E-SSpred, the support vector machine (SVM) method is adopted to model and predict the ends of helices and strands according to the unique residue distributions around them. A simple linear discriminate analysis method is applied to model and predict entire secondary structure segments by integrating end-prediction results, tri-peptide composition, and length distribution features of secondary structures, as well as the prediction results of the most famous program PSIPRED. The results of fivefold cross-validation on a widely used data set demonstrate that the accuracy of E-SSpred in predicting ends of secondary structures is about 10% higher than PSIPRED, and the overall prediction accuracy (Q3 value) of E-SSpred (82.2%) is also better than PSIPRED (80.3%). The E-SSpred web server is available at http://bioinfo.hust.edu.cn/bio/tools/E-SSpred/index.html.
Keywords: secondary structure prediction, position-specific residue preference, ends of secondary structures, protein structure prediction
The knowledge of protein structures plays an important role in understanding protein functions (Watson et al. 2005), reconstructing protein structures (Dwyer et al. 2004), studying protein–protein interactions (Russell et al. 2004), and rationally designing drugs (Thiel 2004). Recently, the gap between available protein sequences and the experimental determination of their structures increased rapidly, making the prediction of protein structures more and more important (Koehl and Levitt 1999; Dunbrack 1999; Baker and Sali 2001). Accurate prediction of protein secondary structures can provide constraints for or be part of a tertiary structure prediction (Russell et al. 1996; Rost 1997; Jones 1999a). Furthermore, knowledge of secondary structures alone can also help the design of site-directed mutants that will not destroy the native protein structures (Chasman and Adams 2001; Bao and Cui 2005).
Secondary structure predictions methods have been developing for many years. The early methods were based on simple statistics (Chou and Fasman 1974; Lim 1974) or stereochemistry principles (Garnier et al. 1978). Thereafter, Qian and his coworkers used the neural network to take the influence of local interactions on secondary structure formation into account, which effectively improved the prediction accuracy (Qian and Sejnowski 1988). In the early 1990s, Rost and Sander (1993) proposed the method of using a sequence profile constructed by a similar sequence search and multiple sequence alignment to predict secondary structures, which exploited the evolution information and improved the prediction accuracy significantly. Later, based on Rost's method (Rost and Sander 1993), Jones (1999b) used PSI-BLAST to improve the homology sequence search, and developed a famous tool named PSIPRED that can get better results.
Today, almost all secondary structure prediction methods follow the Rost's idea (Rost and Sander 1993). These methods build models to predict the secondary structure class of a single residue position according to the information of its neighboring residues (Hua and Sun 2001; Kim and Park 2003; Guo et al. 2004; Qin et al. 2005). Apparently, these methods treat different positions on a protein sequence equally since they predict the secondary structure class of each residue position with the same models. That is, these methods assume that the residue distributions are distinctive for different secondary structure classes, but nondistinctive for different positions of a specific secondary structure class. In fact, the residue distributions on some positions of regular secondary structures are of specificity, which is especially obvious for positions around the ends of regular secondary structures and can be proved by the concept of helix capping (Presta and Rose 1988; Richardson and Richardson 1988; Padmanabhan et al. 1990; Blader et al. 1993; Aurora et al. 1994). Some researchers even declared that the helix ends are determined by the residues around them (Baldwin and Rose 1999). Unfortunately, this position-specific residue preference feature has not been exploited to predict secondary structures. As a result, their prediction performance around the ends of regular secondary structures are quite unsatisfactory, which remarkably limits the application of secondary structure prediction results (Russell and Barton 1993; Rost et al. 1994).
In this study, we collected a relatively large data set consisting of 1860 high-resolution, non-homology proteins from PDB, and further analyzed the residue distributions around the ends of regular secondary structures (i.e., α-helix and β-strands). It was found that there exist position-specific residue preferences around the ends of not only helices but also strands. On this basis, we proposed a novel strategy and developed a tool named E-SSpred to predict the secondary structures. This strategy treats the secondary structure as a whole, and builds models to predict entire secondary structure segments, instead of the class of a single residue, by integrating such information as the residue distribution features around ends, the composition and length distribution features of secondary structure segments, and so on. The results of fivefold cross-validation on the widely used data set CB513 (Cuff and Barton 1999) demonstrate that the accuracy of E-SSpred in predicting ends of secondary structures is about 10% higher than PSIPRED, and the whole prediction accuracy (Q 3 value) of E-SSpred (82.2%) is also better than PSIPRED (80.3%).
Results
Position-specific residue preference around the ends of helices and strands
Based on the DB1860 data set, we analyzed the residue distribution on positions around the ends of helices and strands. Similar to Aurora and coworkers (Aurora et al. 1994), the nomenclatures for these positions are labeled as follows:
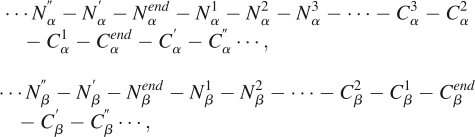 |
where Nendα ,Cendα represent the N-terminal and C-terminal of the α-helices, respectively, and Nendβ and Cendβ represent the N-terminal and C-terminal of the β- strands.
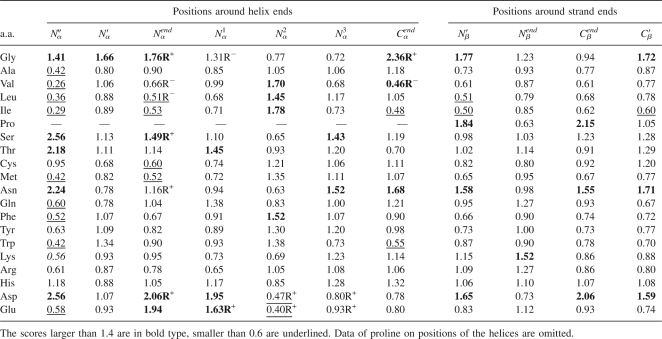
We calculated the residue preference scores (see Equation 1 in Materials and Methods) for each of theses positions. The results for partial positions are given in Table 1. For the purpose of comparison, Richardson's position-specific residue preference results around helix ends (Richardson and Richardson 1988) are also listed in Table 1 (R+ denotes that a residue appears on a position with high frequency, and R− means that the frequency is low). From Table 1, it can be found that many positions exhibit strong position-specific residue preference. For example, on the N-terminal of the helices, the hydrophobic residues such as Val, Leu, and Ile appear infrequently, and the electronegative, polar residues like Asp and Glu, are more likely to present.
Table 1.
Residue preference scores for partial positions around the ends of helices and strands
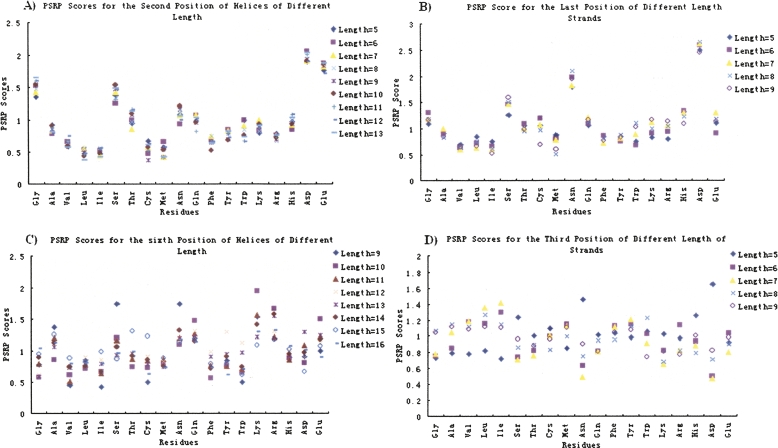
In order to find out whether the residue distributions are influenced by the length of the secondary structures, we further calculated and compared the position-specific residue preference scores for secondary structures of different lengths. Shown in Figure 1 are the results for four selected positions.
Figure 1.
Position-specific residue preference scores for some positions of secondary structures of different lengths. (A) The second position of helices, helix lengths from 5 to 13; (B) the last position of β-strands, strand lengths from 5 to 9; (C) the sixth position of helices, helix lengths from 9 to 16; (D) the third position of strands, stand lengths from 5 to 9. Data of proline are omitted.
From Figure 1 it can be seen that the residue preference scores for the position N 1 α (Fig. 1A) and Cendβ (Fig. 1B) are almost not varying with the length of the secondary structures. It implies that the residue distributions on positions close to the ends of the helices and strands are scarcely influenced by the length of the secondary structures. On the contrary, the residue preference scores for the sixth position of helices (Fig. 1C) and the third position of stands (Fig. 1D), both of them are relatively far away from the ends of secondary structures, greatly vary with the structure length. The results suggest that it is feasible to build a unified model to predict the ends of helices and strands of different lengths.
Accuracy of secondary structure prediction
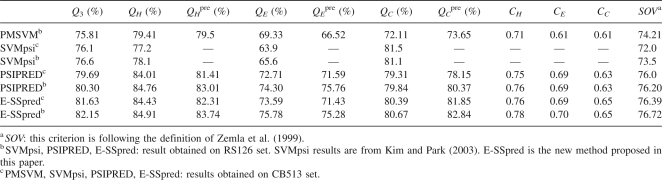
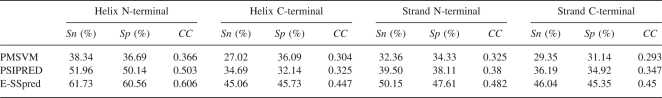
Fivefold cross-validation has been used on RS126 and CB513 to test the performance of E-SSpred, and the results are given in Tables 2 and 3. For the purpose of comparison, the prediction performance of the PMSVM (Guo et al. 2004), SVMpsi (Kim and Park 2003), and PSIPRED (Jones 1999b) on the same data sets, are also given in these tables. In Table 2, three kinds of widely used measures, the per-residue accuracy for overall proteins (Q3 value) and for each class of secondary structure (QH, QE, QC, QH pre, QE pre, Qc pre), Matthew correlation coefficient for each class of secondary structure (CH, CE, CC) (Matthews 1975), and segment overlap measure score (SOV) (Zemla et al. 1999) are used to evaluate the prediction results. The details for calculating per-residue accuracy Q3, QI, and QI pre, Matthew correlation coefficient CI (here, I = H, E, and C), and the segment overlap measure score SOV are given in a previous paper (Kim and Park 2003). In Table 3, three measures, the sensitivity Sn, specificity Sp, and Matthew correlation coefficient CC, are used to evaluate the performance for predicting the ends of helices and strands. The sensitivity is defined as Sn = TP/(TP + FN), the specificity is Sp = TP/(TP + FP), and the Matthews correlation coefficient CC is
Table 2.
The prediction performance of E-SSpred and the comparison with PMSVM, SVMpsi, and PSIPRED
Table 3.
The prediction performance of E-SSpred for the ends of helices and strands and the comparison with PMSVM and PSIPRED
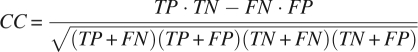 |
where the symbols TP, TN, FP, and FN denote the number of true positives, true negatives, false positives, and false negatives, respectively.
It can be seen from Table 2 that the results from the E-SSpred method are very good. On both data sets of RS126 and CB513, the Q3 value of E-SSpred is improved >5% compared with the recently developed tools PMSVM and SVMpsi, and compared with PSIPRED, one of the most popular secondary structure prediction tools, E-SSpred can also get better performance in terms of the Q3 (increased 2%), correlation coefficient, and SOV value.
The data in Table 3 demonstrate that the performances of both PMSVM and PSIPRED for the prediction of secondary structure ends are quite low. For example, the prediction sensitivity and specificity for helix C-terminal, strand N-terminal, and C-terminal are all under 40%, implying that these tools are incompetent to locate secondary structures exactly. Compared with PMSVM and PSIPRED, the performance of E-SSpred for locating the ends of secondary structures is significantly better. It can be seen from Table 3 that the overall performance (CC) of E-SSpred for predicting helix N-terminal, helix C-terminal, strand N-terminal, and strand C-terminal are higher than PSIPRED, respectively.
Discussion
Position-specific residue preference around the ends of helices and strands
The residue distribution around the ends of helices has been analyzed by Richardson and Richardson (1988) using a small data set containing 45 protein sequences. It was found that, on certain positions, some residues present with preference (e.g., Asp on Nendα, Gly on Cendα, etc.), and some others are unlikely to occur (e.g., Leu on Nendα, Val on Cendα, etc.). In this study, we collected a large data set containing 1860 high-resolution, non-homology proteins to further analyze the residue distributions around the ends of regular secondary structures. It was found that there exist more position-specific residue preferences around the ends of not only helices but also strands (see Table 1). For example, Glu also presents high frequently on position Nendα as Asp, and Asp also prefers to occur on position N 1 α as Glu.
From our results, some interesting conclusions about the residue distribution around the ends of helices and strands can be obtained. For instance, polar and electronegative residues Glu and Asp prefer to present on the first (Nendα) and second positions (N 1 α) of α-helices, but hydrophobic residues such as Val, Leu, Ile, and Met are unlikely to appear on these positions. On the third position of the α-helices (N 2 α), however, hydrophobic residues are of preference but electronegative residues are unlikely to occur. This residue preference on positions next to the helix starts may be one of the requirements to form the helix structure.
Our results show that the position-specific residue preference around the ends of helices and strands is more obvious than the inner positions, which is consistent with the results of Richardson and Richardson (1988). In addition, we also analyzed the residue distributions of secondary structures with different length, and found that the influences of structure length on residue distributions for positions around the ends of helices and strands are much less than those inner positions. These results imply that it is necessary to build specific models to predict the end positions of regular secondary structures.
There are also some conflicts between Richardson and Richardson's (1988) results and ours. For example, their research showed that both Asp and Glu prefer to present on N 2 α, but our results indicate that these two residues are unlikely to present on N 2 α (Richardson and Richardson 1988). The main cause for these conflicts is possibly that too few protein sequences were used in Richardson and Richardson's (1988) research to estimate the residue distributions, which might lead to some statistical biases.
The performance of secondary structure prediction
From Table 3 it can be seen that the performance of E-SSpred for predicting the ends of α-helices and β-strands is significantly better than PMSVM, SVMpsi, and PSIPRED, indicating that the position-specific residue preference around the ends of helices and strands is a very useful feature to help predict the ends of secondary structures and locate secondary structures more accurately. Moreover, it means that E-SSpred can locate the secondary structures on protein sequences more accurately, and therefore its prediction results can be applied to solve related problems such as protein tertiary structure prediction, protein function analysis, and so on, more effectively. However, from Table 2, it can also be seen that, with the help of this feature, the improvement of secondary structure prediction performance in terms of the measures Q3, Matthew correlation coefficient, and SOV is not very significant. The possible causes include: (1) The number of ends is very small relative to the number of residues in helices and strands; thus, the direct contribution of improving the prediction of ends is limited to the improvement of secondary structure prediction accuracy measured by Q3, etc.; (2) the main cause that greatly influences the performance of existing secondary structure prediction tools is that some helices and strands are easily predicted as loops completely. The novel strategy proposed in this study, to predict entire secondary structure segments directly by integrating relevant features in such aspects as the residue distribution around ends, tri-peptide composition, and so on, has the potential to change this situation. However, the algorithms currently used in E-SSpred are still too simple to adequately bring into play the potential of this novel strategy. We expect to develop, in the near future, more advanced algorithms that can significantly improve the prediction performance.
Materials and Methods
Data sets
Three data sets were used in this study. One is the data set we collected to analyze the statistical features of secondary structures and to train models for secondary structure ends prediction. This data set contains 1860 non-homology proteins and is called DB1860. The proteins in this data set were picked from the PDB database using the tool, PISCES, developed by Dunbrack (Wang and Dunbrack 2003). These proteins meet the following criterions: (1) they were detected by an X-ray diffraction method; (2) the sequence identity between any two of them is <30%; (3) the experiment resolution is <2.0 angstroms; (4) there are no homology proteins between DB1860 with RS126 and CB513 data sets. The list of proteins in DB1860 can be downloaded from the website: http://bioinfo.hust.edu.cn/bio/tools/E-SSpred/.
The other two data sets are RS126 (constructed by Rost and Sander [1993]) and CB513 (constructed by Cuff and Barton [1999]); these two data sets contains 126 and 513 non-homology proteins, respectively, and have been widely used to test secondary structure prediction methods (Hua and Sun 2001; Kim and Park 2003), and they also are used to compare the prediction performance of our method with that of other methods.
The secondary structure of proteins in these data sets is assigned from the experimentally determined tertiary structure by DSSP (Kabsch and Sander 1983), which has been the most widely used secondary structure definition. It has eight secondary structure classes: H(α-helix), G(310-helix), I(π-helix), E(β-strand), B(isolated β-bridge), T(turn), S(bend), and –(rest). We reduced the eight classes to three states, helix(H), sheet(E), and coil(C) using the following strategy: H, G to H; E, B to E; all other states to C. This strategy is now widely used, and considered to be the strictest definition in secondary structure prediction methods (Hua and Sun 2001; Kim and Park 2003; Guo et al. 2004).
Assessment of position-specific residue preference around ends of secondary structures
The position-specific residue preference is defined as the statistical frequencies where residues occur on a certain position around the ends of secondary structures. The preference score for residue a on position i of secondary structure class ss is denoted as fss (a, i), and is determined by:
where pss (a, i) is the frequency of residue a occurring on position i of secondary structure class ss, and p 0 ss (a) is the average frequency of residue a on all positions of ss.
The position-specific residue preference for secondary structures of different lengths is denoted as flss (a, i), and is determined by
where l is the length of secondary structures, plss (a, i) is the frequency of residues a occurring on position i of structure ss whose length is l, and pl 0 ss (a) is the average frequency.
Prediction of ends of secondary structures
We first predict the probabilities of each position in the protein belonging to the end positions of regular secondary structures. The SVM method is adopted to do this job according to the residue distributions around each position. For each kind of ends, the helix N-terminal, helix C-terminal, strand N-terminal, and strand C-terminal, a binary SVM classifier, is built to predict them, respectively. By the analysis of the position-specific residue preference scores of different positions in secondary structures, we find that the position-specific residue preference features on some positions, such as nine positions around the helix N-terminal (i.e., upstream three residues, downstream five residues, and the helix N-terminal itself), and seven positions around other terminals (i.e., upstream three residues, downstream three residues, and the end itself), is more intense than on other positions. Based on this, to predict the helix N-terminal, nine residues are encoded with PSSM (position-specific scores matrix) scores to construct feature vectors, and for the prediction of the other three ends, seven residues are selected to construct feature vectors.
In this study, the LIBSVM (http://www.csie.ntu.edu.tw/∼cjlin/libsvm/) program is used as the implementation of SVM, in which the radial basis function kernel is adopted and the two parameters, C and γ, are empirically set to 10 and 0.0015, respectively.
In the course of prediction, for each position of a protein sequence, these SVMs can output four scores reflecting the probabilities of this position being a helix N-terminal, helix C-terminal, strand N-terminal, and strand C-terminal, respectively, and these scores will be used as features in the secondary structure prediction of the whole protein.
Tri-peptide composition in secondary structures
Similar to the idea of using codon usage to help distinguish exons from introns in the field of predicting gene structures in eukaryotic DNA sequences, in this study, the tri-peptide composition is used as an additional feature to help distinguish different secondary structures. For a tri-peptide, its probability score, appearing in secondary structure ss, is defined as:
where ai, aj, ak denote a residue type, respectively, n(aiajak | ss) is the number that the tri-peptide aiajak appears as in ss.
Length distribution of secondary structures
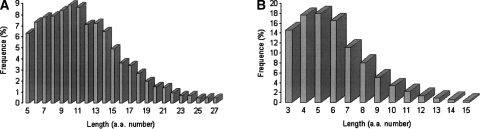
As shown in Figure 2, the length distribution of helices is different from that of strands. Thus, the length can be used as an additional feature to help distinguish different secondary structures. In this study, the length score for predicting a segment of length l as a helix or strand is determined by:
Figure 2.
The length distribution of (A) helices and (B) strands in DB1860.
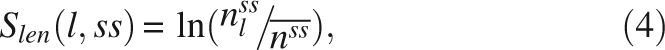 |
where nssl is the number of secondary structures of length l of secondary structures of class ss, and 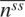 is the average number of all lengths of ss. Both nssl and
is the average number of all lengths of ss. Both nssl and 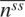 are determined by the training data set DB1860.
are determined by the training data set DB1860.
Linear discriminate analysis of secondary structures
Similar to the strategy widely used in predicting exons and introns of genes from DNA sequences, in this study, the linear discriminate analysis method is used to integrate the end-prediction results, tri-peptide composition score, length distribution score, and the PSIPRED prediction results to predict entire secondary structure segments.
For a protein sequence, let Seg[i, j] be a segment of this sequence (i and j are the start and end position, respectively), then the potential score where this segment belongs to helices or strands is determined by:
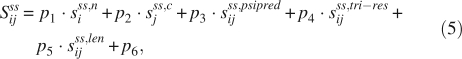 |
where, sss,ni is the probability score for position i to be the N-terminal of class ss, sss,cj is the score for position j to be the C-terminal of ss, sss,lenij and sss,tri–resij are the length distribution score and average tri-peptide composition score of this segment, respectively. sss,psipredij is the PSIPRED prediction score determined by
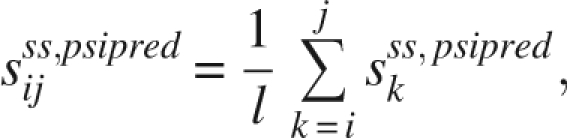 |
and the sss,psipredk is the score of residues k predicted to be ss by PSIPRED. The parameters p 1 – p 6 are weight coefficients that are determined by the least-square approach used for the REGRESS function of Matlab7.0.
A segment whose potential score Sssij is bigger than a threshold Sssthreshold is predicted as an ss candidate, and all the candidates in a protein sequence can be obtained after scanning the whole sequence. For overlapped candidates of the same class, only the one with the biggest Sssij value is kept. Then, for overlapped candidates of different classes (for example, a helix candidate from position i to j, a strand candidate from k to l, and i < k < j < l), the following rules are used to make the decision:
where H and E represent the helix and the strand, respectively, and struct(i → j) = H means the segment from position i to j is predicted as a helix and so do the others.
Acknowledgments
We thank J.A. Cuff and G.J. Barton for providing the CB513 data set. This work was supported by the National Natural Science Foundation of China (Grant Nos. 90608020, 30370354, and 90203011), NCET-060651, the National Platform Project of China (Grant No. 2005DKA64001), and the Ministry of Education of China (Grant Nos. 20050487037 and 505010).
Footnotes
Reprint requests to: Yanhong Zhou, Hubei Bioinformatics and Molecular Imaging Key Laboratory, Huazhong University of Science and Technology, Wuhan 430074, People's Republic of China; e-mail: yhzhou@hust.edu.cn; fax: 86-27-87792170.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.035691.108.
References
- Aurora, R., Srinivasan, R., Rose, G.D. Rules for α-helix termination by glycine. Science. 1994;264:1126–1130. doi: 10.1126/science.8178170. [DOI] [PubMed] [Google Scholar]
- Baker, D., Sali, A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- Baldwin, R.L., Rose, G.D. Is protein folding hierarchic? I. Local structure and peptide folding. Trends Biochem. Sci. 1999;24:26–33. doi: 10.1016/s0968-0004(98)01346-2. [DOI] [PubMed] [Google Scholar]
- Bao, L., Cui, Y. Prediction of the phenotypic effects of non-synonymous single nucleotide polymorphisms using structural and evolutionary information. Bioinformatics. 2005;21:2185–2190. doi: 10.1093/bioinformatics/bti365. [DOI] [PubMed] [Google Scholar]
- Blader, M., Zhang, X.J., Matthews, B.W. Structural basis of amino acid α-helix propensity. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- Chasman, D., Adams, R.M. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: Structure-based assessment of amino acid variation. J. Mol. Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- Chou, P.Y., Fasman, G.D. Conformational parameters for amino acids in helical, -sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Cuff, J.A., Barton, G.J. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Dunbrack, R.L. Sequence comparison and protein structure prediction. Curr. Opin. Struct. Biol. 1999;16:374–384. doi: 10.1016/j.sbi.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Dwyer, M.A., Looger, L.L., Hellinga, H.W. Computational design of a biologically active enzyme. Science. 2004;304:1967–1971. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- Garnier, J., Osguthorpe, D.J., Robson, B. Analysis and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Guo, J., Chen, H., Sun, Z.R., Lin, Y.L. A novel method for protein secondary structure prediction using dual-layer SVM and profiles. Proteins. 2004;54:738–743. doi: 10.1002/prot.10634. [DOI] [PubMed] [Google Scholar]
- Hua, S.J., Sun, Z.R. A novel method of protein secondary structure prediction with high segment overlap measure: Support vector machine approach. J. Mol. Biol. 2001;308:397–407. doi: 10.1006/jmbi.2001.4580. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. GenTHREADER: An efficient and reliable protein folding recognition method for genomic sequences. J. Mol. Biol. 1999a;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. Protein secondary structure prediction based on position-specific score matrix. J. Mol. Biol. 1999b;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., Sander, C. Dictionary of protein secondary structure: Pattern recognition of hudrogen bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kim, H., Park, H. Protein secondary structure prediction based on an improved support vector machines approach. Protein Eng. 2003;16:553–560. doi: 10.1093/protein/gzg072. [DOI] [PubMed] [Google Scholar]
- Koehl, P., Levitt, M. A bright future for protein structure prediction. Nat. Struct. Biol. 1999;6:108–111. doi: 10.1038/5794. [DOI] [PubMed] [Google Scholar]
- Lim, V.I. Algorithms for prediction of α-helices and structural regions in globular proteins. J. Mol. Biol. 1974;88:872–894. doi: 10.1016/0022-2836(74)90405-7. [DOI] [PubMed] [Google Scholar]
- Matthews, B.W. Comparison of the prediction and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, S., Marquesee, S., Ridgeway, T., Laue, T.M., Baldwin, R.L. Relative helix-forming tendencies of nonpolar amino acids. Nature. 1990;344:268–270. doi: 10.1038/344268a0. [DOI] [PubMed] [Google Scholar]
- Presta, L.G., Rose, G.D. Helix signals in proteins. Science. 1988;240:1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- Qian, N., Sejnowski, T.J. Predicting the neural network models. J. Mol. Biol. 1988;202:865–884. doi: 10.1016/0022-2836(88)90564-5. [DOI] [PubMed] [Google Scholar]
- Qin, S.B., He, Y., Pan, X.M. Predicting protein secondary structure and solvent accessibility with an improved multiple linear regression method. Proteins. 2005;61:473–480. doi: 10.1002/prot.20645. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S., Richardson, D.C. Amino acid preference for specific locations at the ends of helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Rost, B. Protein fold recognition by prediction based threading. J. Mol. Biol. 1997;270:1–10. doi: 10.1006/jmbi.1997.1101. [DOI] [PubMed] [Google Scholar]
- Rost, B., Sander, C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Rost, B., Sander, C., Schneider, R. Redefining the goals of protein secondary structure prediction. J. Mol. Biol. 1994;235:13–26. doi: 10.1016/s0022-2836(05)80007-5. [DOI] [PubMed] [Google Scholar]
- Russell, R.B., Barton, G.J. The limits of protein secondary structure prediction accuracy from multiple sequence alignment. J. Mol. Biol. 1993;234:951–957. doi: 10.1006/jmbi.1993.1649. [DOI] [PubMed] [Google Scholar]
- Russell, R.B., Copley, R.R., Barton, G.J. Protein fold recognition by mapping predicted secondary structures. J. Mol. Biol. 1996;259:349–365. doi: 10.1006/jmbi.1996.0325. [DOI] [PubMed] [Google Scholar]
- Russell, R.B., Alber, F., Aloy, P., Davis, F.P., Korkin, D., Pochaud, M., Topf, M., Sali, A. A structural perspective on protein–protein interactions. Curr. Opin. Struct. Biol. 2004;14:313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Thiel, K.A. Structure-aided drug design's next generation. Nat. Biotechnol. 2004;22:513–519. doi: 10.1038/nbt0504-513. [DOI] [PubMed] [Google Scholar]
- Wang, G., Dunbrack, J.R. PISCES: A protein sequence-culling serve. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- Watson, J.D., Laskowski, R.A., Thornton, J.M. Predicting protein function from sequence and structural data. Curr. Opin. Struct. Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Zemla, A., Venclovas, C., Fidelis, K., Rost, B. A modified definition of Sov, a segment-based measure for protein secondary structure prediction assessment. Proteins. 1999;34:220–223. doi: 10.1002/(sici)1097-0134(19990201)34:2<220::aid-prot7>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]