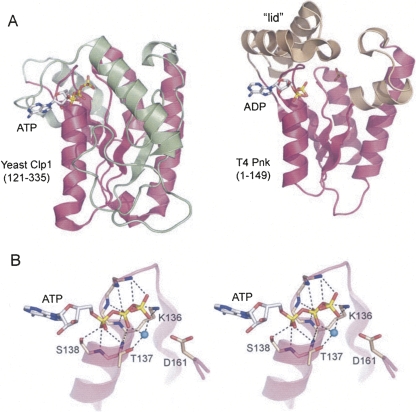
FIGURE 2.
Structural homology between Clp1 and T4 Pnk. (A) The structures of the yClp1 phosphoesterase domain (PDB ID 2NPI) and the T4 Pnk kinase domain (PDB ID 1LTQ and 1LY1) were superimposed and then offset horizontally. The folds are shown as ribbon traces with shared structural elements (magenta). (Green) Distinctive structural components of yClp1, (beige) distinctive structural components of T4 Pnk. The ATP and ADP ligands are depicted as stick models. In the T4 Pnk structure, a sulfate ion occupies the position of the terminal phosphodiester of the 5′-OH polynucleotide substrate. (B) Stereo view of the ATP-binding site of yClp1 highlighting the oxyanion hole formed by main-chain amides and the side chains of the P-loop motif QTGKTS138. (Cyan sphere) Magnesium ion, (dashed lines) atomic contacts of yClp1 with ATP and magnesium.