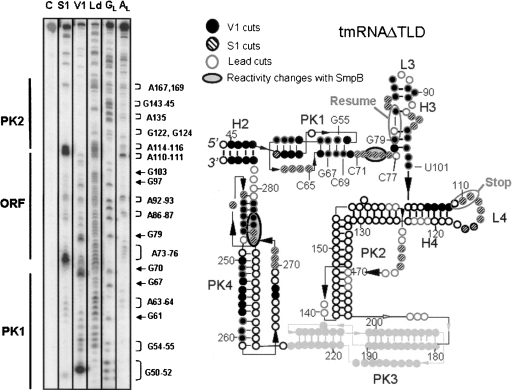
FIGURE 3.
Probing the conformation of tmRNAΔTLD in solution. (Left) Autoradiogram of cleavage products of 5′-labeled tmRNAΔTLD by lead acetate (Ld), nuclease S1, and RNase V1. Lane C, incubation control; lanes GL and AL, RNases T1 and U2 hydrolysis ladders, respectively. The RNA sequence is indexed on the right and the structural domains on the left of the panel. (Right) Secondary structure of tmRNAΔTLD from A. aeolicus inferred by structural mapping in solution. The numbering corresponds to that of full-length tmRNA, for direct comparison. The meaning of the symbols is detailed in the inset. Structural domains, such as the pseudoknots (PK), the helices (H), and the loops (L), are indicated. The internal open reading frame (ORF) is indicated. Structural information is missing for PK3 (pale gray). The black bars are the Watson–Crick (GC or AU) pairs. The resume and stop codons are circled.