Figure 5.
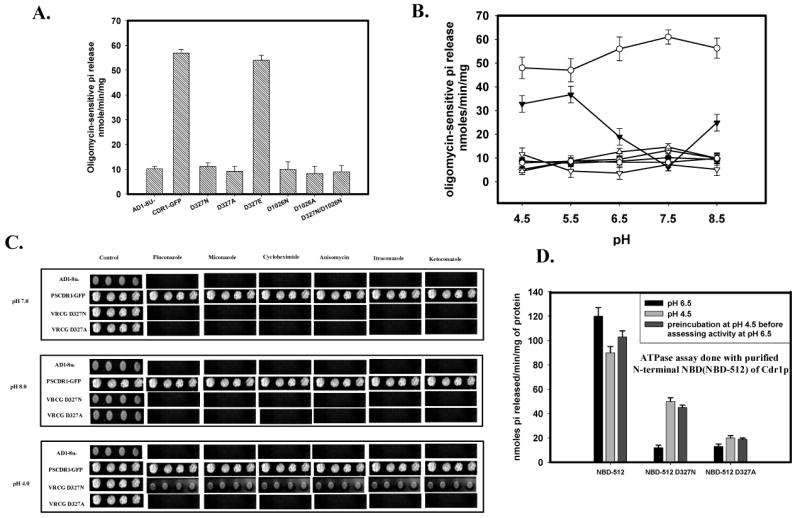
(A) ATPase activity of Cdr1p with its mutant variants: ATPase activity of PM fraction of cells expressing the wild type Cdr1p and its mutant variants were assayed as described previously (26). The values were the mean ± standard deviation of three independent experiments. (B) pH dependence of ATPase activity of Cdr1p and its mutant variants: ATPase activity of PM fractions expressing wild type Cdr1p and its mutant variants were assayed at different pH. Scattered plot represents AD1-8u- (●) PSCdr1-GFP (○), VRCG D327N (▼), VRCG D327A (▽), VRCG D1026N (◊), VRCG D1026A (
 ) and VRCG D327N/D1026N (Δ). (C) Reversal of drug resistance at low pH by VRCG D327N strain: Reversal of drug resistance by VRCG D327N strain at different pH was corroborated by spot assay by growing the cells at different pH in presence of drug; Flu (0.5 μg/ml), Mico (0.125 μg/ml), Keto (0.0156 μg/ml), Itra (0.125 μg/ml), Cyclo (0.0625μg/ml) and Aniso (0.5 μg/ml). (D) ATPase activity of isolated N-terminal NBD (NBD-512) of Cdr1p and its mutant variant proteins: ATPase activity of NBD-512 and its mutant variant proteins at pH 6.5 was assayed after pre-incubation of the protein at low pH (4.5).
) and VRCG D327N/D1026N (Δ). (C) Reversal of drug resistance at low pH by VRCG D327N strain: Reversal of drug resistance by VRCG D327N strain at different pH was corroborated by spot assay by growing the cells at different pH in presence of drug; Flu (0.5 μg/ml), Mico (0.125 μg/ml), Keto (0.0156 μg/ml), Itra (0.125 μg/ml), Cyclo (0.0625μg/ml) and Aniso (0.5 μg/ml). (D) ATPase activity of isolated N-terminal NBD (NBD-512) of Cdr1p and its mutant variant proteins: ATPase activity of NBD-512 and its mutant variant proteins at pH 6.5 was assayed after pre-incubation of the protein at low pH (4.5).
