Abstract
The cytokine interleukin (IL)-1β is a key mediator of the inflammatory response and has been implicated in the pathophysiology of acute and chronic inflammation. IL-1β is synthesized in response to many stimuli as an inactive pro–IL-1β precursor protein that is further processed by caspase-1 into mature IL-1β, which is the secreted biologically active form of the cytokine. Although stimulation of membrane-bound Toll-like receptors (TLRs) up-regulates pro–IL-1β expression, activation of caspase-1 is believed to be mainly initiated by cytosolic Nod-like receptors. In this study, we show that polyinosinic:polycytidylic acid (poly[I:C]) and lipopolysaccharide stimulation of macrophages induces pro–IL-1β processing via a Toll/IL-1R domain–containing adaptor-inducing interferon-β–dependent signaling pathway that is initiated by TLR3 and TLR4, respectively. Ribonucleic acid interference (RNAi)–mediated knockdown of the intracellular receptors NALP3 or MDA5 did not affect poly(I:C)-induced pro–IL-1β processing. Surprisingly, poly(I:C)- and LPS-induced pro–IL-1β processing still occurred in caspase-1–deficient cells. In contrast, pro–IL-1β processing was inhibited by caspase-8 peptide inhibitors, CrmA or vFLIP expression, and caspase-8 knockdown via RNAi, indicating an essential role for caspase-8. Moreover, recombinant caspase-8 was able to cleave pro–IL-1β in vitro at exactly the same site as caspase-1. These results implicate a novel role for caspase-8 in the production of biologically active IL-1β in response to TLR3 and TLR4 stimulation.
IL-1β is a master cytokine that mediates several immune responses and is synthesized as an inactive precursor that is processed into biologically active IL-1β in response to various proinflammatory stimuli (1). It is generally accepted that pro–IL-1β processing in response to infection and other proinflammatory conditions is mediated by caspase-1 (2). There are 11 caspases in humans, but only caspase-1 has been shown to mediate pro–IL-1β processing. Many caspases are implicated in apoptosis, but certain caspases also exert nonapoptotic functions, including proliferation, differentiation, and NF-κB activation (3).
Recognition of Toll-like receptors (TLRs) by microbial or other danger-associated molecules induces the NF-κB–dependent transcription of the IL-1β gene encoding an inactive pro–IL-1β protein. Signaling leading to the proteolytic processing of pro–IL-1β by caspase-1 is initiated by a distinct set of so-called Nod-like receptors (NLRs) as part of the “inflammasome,” which is an intracellular multiprotein complex that also contains caspase-1 (2, 4–10). In this study, we demonstrate the existence of a Toll/IL-1R domain–containing adaptor-inducing IFN-β (TRIF)–dependent signaling pathway that mediates processing and secretion of IL-1β in response to TLR3 and TLR4 stimulation. Most interestingly, we show that TLR3- and TLR4-induced pro–IL-1β processing is mediated by caspase-8.
RESULTS AND DISCUSSION
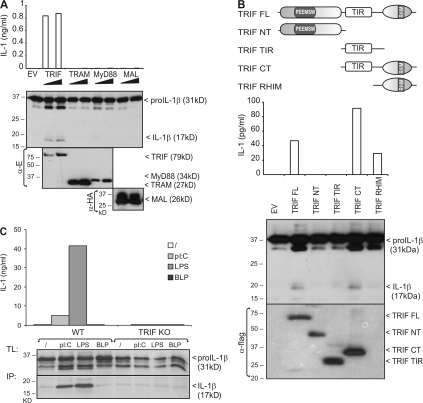
We first examined the potential of TLRs to initiate pro–IL-1β processing. TLR signaling depends on four different adaptor proteins (MyD88, MAL/TIRAP, TRAM/TICAM-2, and TRIF/TICAM-1), which bind to specific TLRs and mediate two main signaling pathways, leading to activation of NF-κB and IFN regulatory factor (IRF) transcription factors (11). The LPS receptor TLR4 uses MAL and TRAM as bridging adaptors for MyD88 and TRIF, respectively. The double-stranded RNA receptor TLR3 only needs TRIF, whereas all other TLRs signal via MyD88. TLR2 also requires MAL to recruit MyD88. Overexpression of each TLR adaptor was previously shown to activate NF-κB. Therefore, in a similar approach, we first tested whether overexpression of specific TLR adaptor proteins in human embryonic kidney 293T (HEK293T) cells triggers processing and secretion of ectopically expressed pro–IL-1β. Production of mature IL-1β was measured in an IL-1 bioassay (Fig. 1 A, top), as well as by Western blotting (Fig. 1 A, bottom). Interestingly, whereas all four TLR adaptors induced the activation of an NF-κB–dependent reporter gene (unpublished data), mature IL-1β production could only be detected upon overexpression of the TLR3 and TLR4 adaptor protein TRIF. TRIF signaling to NF-κB is known to involve the binding of the TRIF N-terminal domain with TRAF6, as well as the binding of the TRIF C-terminal receptor–interacting protein (RIP) homology interaction motif (RHIM) with RIP1 (12, 13). Deletion of the C-terminal Toll/IL-1 receptor domain (TIR) and RHIM containing part of TRIF completely abolished its ability to induce pro–IL-1β maturation (Fig. 1 B). On the other hand, a TRIF mutant lacking the TIR domain, but still containing the more C-terminal RHIM domain, was equally potent as full-length TRIF. These data illustrate an important role of the C-terminal RHIM containing domain of TRIF in signaling to pro–IL-1β processing.
Figure 1.
Poly(I:C) and LPS induce pro–IL-1β processing via a TRIF-dependent signaling pathway. (A) HEK293T cells were cotransfected with pro–IL-1β and 50 or 100 ng of either E-TRIF, E-TRAM, E-MyD88, or HA-MAL. 24 h later, pro–IL-1β processing and expression of transfected proteins was analyzed by Western blotting of total cell lysates (bottom). Secretion of biologically active IL-1β into the corresponding cell supernatants was analyzed via IL-1 bioassay (top). (B) HEK293T cells were cotransfected with pro–IL-1β and different Flag-tagged TRIF deletion mutants. 24 h later, pro–IL-1β processing and secretion of biologically active IL-1β was analyzed as in A. Expression of TRIF mutants was verified by Western blotting and detection with anti-Flag. (top) Schematic representation of the different TRIF deletion mutants. FL, full length; NT, N-terminal fragment; CT, C-terminal fragment; PEEMSW, TRAF6-binding motif; TIR, Toll/IL-1 receptor domain. (C) BLP-primed peritoneal macrophages from WT and TRIF KO mice were incubated for 18 h with poly(I:C), LPS, BLP, or control medium, as described in the Materials and methods. Pro–IL-1β was analyzed in total cell lysates (TL) by Western blotting (bottom). Mature IL-1β was detected in the cell supernatant upon IL-1β immunoprecipitation (IP). Secretion of biologically active IL-1β was analyzed via IL-1 bioassay (top). Data are representative of three independent experiments.
The specific ability of TRIF overexpression to trigger pro–IL-1β processing indicates that TRIF might mediate pro–IL-1β processing in response to TLR3 and TLR4 stimulation. We therefore tested the ability of polyinosinic:polycytidylic acid (poly[I:C]) and LPS, respectively, to initiate pro–IL-1β processing in either WT or TRIF-deficient peritoneal macrophages. To exclude a contribution of TRIF-induced changes in pro–IL-1β expression caused by the ability of TRIF to also activate NF-κB, cells were stimulated with poly(I:C) and LPS in the presence of the translation inhibitor cycloheximide (CHX), and pro–IL-1β expression was first induced by pretreating the cells with the synthetic bacterial lipoprotein (BLP) and TLR2 ligand Pam3Cys-SK4, which was previously shown to up-regulate pro–IL-1β without inducing its processing (6). Under these conditions, poly(I:C) and LPS significantly induced pro–IL-1β processing and secretion of mature IL-1β by peritoneal macrophages, which was completely prevented in TRIF-deficient macrophages (Fig. 1 C). As expected, BLP did not trigger pro–IL-1β processing. It should be mentioned that CHX not only prevented the up-regulation of pro–IL-1β by LPS and poly(I:C), but also increased the constitutive and inducible processing of pro–IL-1β (unpublished data). A similar effect of CHX has recently been described for pro–IL-1β processing in response to muramyl dipeptide binding to NOD2 (7). This probably reflects the inhibitory effect of CHX on the expression of a negative regulator of pro–IL-1β processing.
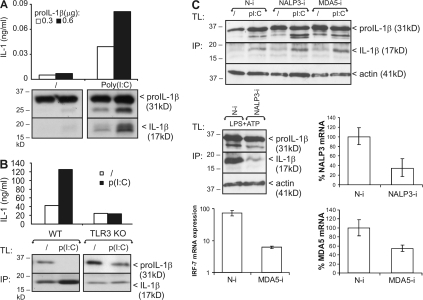
Besides binding to membrane-bound TLR3, poly(I:C) is also recognized by cytosolic MDA5, which triggers NF-κB and IRF activation (14). In addition, NALP3 (also known as Cryopyrin) is a cytosolic NLR that has been shown to play a critical role in the activation of caspase-1 in response to double-stranded RNA treatment of bone marrow–derived macrophages (8). To further demonstrate that the poly(I:C)-induced pro–IL-1β processing that we observe is initiated by TLR3, we tested pro–IL-1β processing in HEK293-TLR3 cells and BLP-primed peritoneal macrophages from WT and TLR3-deficient mice, respectively. Poly(I:C) clearly stimulated pro–IL-1β processing in HEK293-TLR3 cells (Fig. 2 A) and WT macrophages, but not in TLR3-deficient macrophages (Fig. 2 B). Moreover, pro–IL-1β processing after poly(I:C) treatment of BLP-primed macrophages was unaffected by MDA5 and NALP3 small interfering RNA (siRNA; Fig. 2 C, top). Knockdown was verified via quantitative PCR (qPCR) of NALP3 and MDA5 mRNA (Fig. 2 C, middle and bottom right). Decreased pro–IL-1β processing in response to LPS plus ATP, which is known to require NALP3 (9, 10), was used as a positive control for the effect of NALP3 siRNA (Fig. 2 C, middle left). Similarly, decreased IRF-7 mRNA induction upon intracellular delivery of poly(I:C), which is known to initiate MDA5 signaling to type I IFN production, leading to IRF-7 expression (14), was used as a positive control for the effect of MDA5 siRNA (Fig. 2 C, bottom left). Collectively, these data demonstrate that the stimulatory effect of extracellular poly(I:C) on pro–IL-1β processing is mediated by TLR3 and its adaptor TRIF.
Figure 2.
Poly(I:C)-induced pro–IL-1β processing is TLR3 dependent. (A) HEK293-TLR3 cells transfected with 0.3 or 0.6 μg pCAGGS-pro–IL-1β were incubated in the absence or presence of 25 μg/ml poly(I:C) for 6 h. Pro–IL-1β processing was analyzed by SDS-PAGE and Western blotting of total cell lysates (bottom). Secretion of biologically active IL-1β into the corresponding cell supernatants was analyzed via IL-1 bioassay (top). (B) BLP-primed peritoneal macrophages from WT and TLR3 KO mice were incubated for 18 h with poly(I:C) or control medium, as described in the Materials and methods. Pro–IL-1β processing was analyzed by Western blotting of total cell lysates (TL) and IL-1β immunoprecipitates (IP) from the cell supernatant (bottom). Secretion of biologically active IL-1β was analyzed via IL-1 bioassay (top). (C) Peritoneal macrophages were transfected with either control nontargeting (N-i), NALP3 siRNA (NALP3-i), or MDA5 siRNA (MDA5-i), as indicated. (top) 72 h later, cells were treated and analyzed for pro–IL-1β processing as described in B. (middle right and bottom right) Knockdown was verified by qPCR of NALP3 and MDA5, and is presented as a percentage of the mRNA levels in cells that were not treated with siRNA. As a positive control for the effect of NALP3 siRNA, pro–IL-1β processing was measured in cells that were stimulated for 12 h with 100 ng/ml LPS, pulsed for 20 min with 5 mM ATP, and subsequently incubated in fresh medium for 3 h (middle left). As a positive control for the effect of MDA5 siRNA, IRF-7 mRNA expression was measured via qPCR 6 h after transfecting 5 μg/ml poly(I:C) (bottom left). Data are representative of three independent experiments.
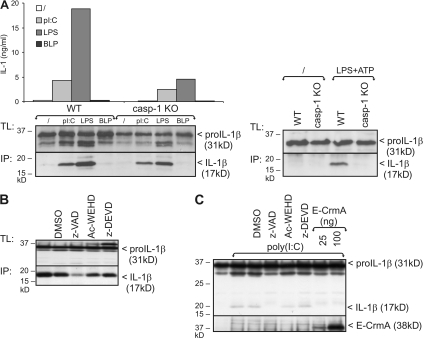
NLR-mediated activation of caspase-1 is believed to be responsible for pro–IL-1β processing in response to multiple stimuli (2, 4–10). To study the role of caspase-1 in the TLR-initiated pro–IL-1β processing observed in this study, we compared the potential of poly(I:C) and LPS to stimulate pro–IL-1β processing in BLP-primed peritoneal macrophages from WT and caspase-1–deficient mice. As a positive control, peritoneal macrophages were stimulated with LPS plus ATP, which induces caspase-1–mediated pro–IL-1β processing by activating the NLR NALP3 (9, 10). Interestingly, poly(I:C)- or LPS-induced pro–IL-1β processing were only slightly reduced in caspase-1–deficient macrophages compared with WT macrophages (Fig. 3 A, left), whereas LPS plus ATP-induced pro–IL-1β processing was completely abolished in the absence of caspase-1 (Fig. 3 A, right). These data indicate that TLR3 and TLR4 can trigger pro–IL-1β processing independent of caspase-1. This was further confirmed by the fact that treatment of peritoneal macrophages or pro–IL-1β transfected HEK293-TLR3 cells with the caspase-1 inhibitor Ac-WEHD-cho fails to completely inhibit poly(I:C)-induced pro–IL-1β processing (Fig. 3, B and C). Remarkably, the pan-caspase inhibitor z-VAD-fmk significantly inhibited pro–IL-1β processing, whereas the caspase-3 and -7 inhibitor z-DEVD-cmk had no effect. In addition, overexpression of CrmA, which is a potent inhibitor of caspase-1, -4, -5, and -8 (15), diminished poly(I:C)-induced pro–IL-1β processing in HEK293-TLR3 cells (Fig. 3 C).
Figure 3.
Poly(I:C) and LPS-induced pro–IL-1β processing is caspase-1 independent. (A) BLP-primed peritoneal macrophages from WT and caspase-1 KO mice were incubated for 18 h with poly(I:C), LPS, BLP, or control medium, as described in the Materials and methods. Pro–IL-1β processing was analyzed by Western blotting of total cell lysates (TL) and IL-1β immunoprecipitates (IP) from the cell supernatant (left). As a positive control for caspase-1–mediated pro–IL-1β processing, cells were stimulated for 12 h with 100 ng/ml LPS, pulsed for 20 min with 5 mM ATP, and subsequently incubated in fresh medium for 3 h (right). (B) Peritoneal macrophages were stimulated for 18 h with poly(I:C), as described in A. 1 h before incubation, cells received 50 μM z-VAD-fmk, Ac-WEHD-cho, z-DEVD-cmk, or 0.05% DMSO (solvent control). Pro–IL-1β processing was analyzed by Western blotting of total cell lysates (TL) and IL-1β immunoprecipitates (IP) from the cell supernatant (bottom). (C) HEK293-TLR3 cells transfected with 0.6 μg pCAGGS-pro–IL-1β were incubated for 6 h with 25 μg/ml poly(I:C). 1 h before incubation, cells received 50 μM z-VAD-fmk, Ac-WEHD-cho, z-DEVD-cmk, or 0.05% DMSO (solvent control). In the last two lanes, HEK293-TLR3 cells were cotransfected with two different concentrations of CrmA-E. Pro–IL-1β processing was analyzed by SDS-PAGE and Western blotting of total cell lysates. Expression of CrmA was verified by Western blotting and anti-E tag. Data are representative of three independent experiments.
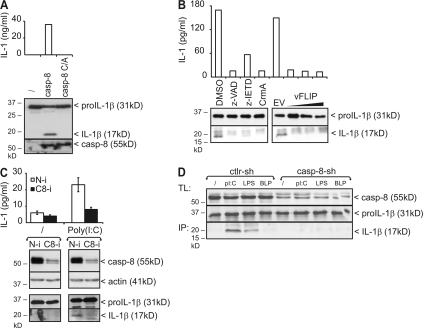
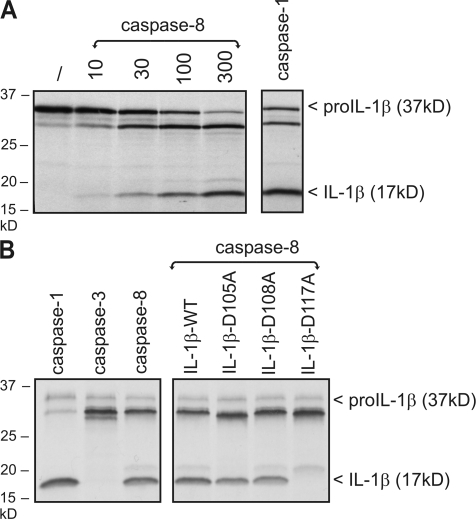
It should be noted that several proteases other than caspase-1 were previously shown to proteolytically activate pro–IL-1β under certain conditions (16). However, none of these belong to the caspase family. Our observation that poly(I:C)-induced pro–IL-1β processing can be inhibited by z-VAD-fmk and CrmA pointed to a role for caspase-8. This was further suggested by the fact that overexpression in HEK293T cells of WT caspase-8, but not its catalytically inactive mutant, induced pro–IL-1β processing and secretion of biological active IL-1β (Fig. 4 A). Similar results were obtained upon overexpression of caspase-1 (unpublished data). Consistent with the aforementioned inhibitory effect of z-VAD-fmk and CrmA, also more specific inhibition of caspase-8 with z-IETD-fmk or the viral caspase-8 inhibitor vFLIP prevented TLR3-induced pro–IL-1β processing (Fig. 4 B). Similarly, siRNA-mediated knockdown of caspase-8 completely prevented poly(I:C)-induced processing in HEK293-TLR3 cells (Fig. 4 C), as well as poly(I:C)- and LPS-induced pro–IL-1β processing in RAW264.7 murine macrophages (Fig. 4 D). Altogether, these experiments demonstrate a crucial role for caspase-8 in the production of mature IL-1β in response to TLR3 and TLR4 stimulation. To analyze whether caspase-8 can directly cleave pro–IL-1β into its mature form, we compared the ability of recombinant caspase-1 and -8 to process pro–IL-1β in vitro. Both caspases were able to process pro–IL-1β into an intermediate and mature form (Fig. 5 A). In contrast, caspase-3 only generated the intermediate form (Fig. 5 B, left). Caspase-1 produces mature IL-1β by cleaving pro–IL-1β after Asp117 (17). Interestingly, mutation of Asp117 also prevented the in vitro generation of mature IL-1β by caspase-8, whereas mutation of two other potential cleavage sites that are conserved in mouse and human pro–IL-1β (Asp105 and Asp108) had no effect (Fig. 5 B). These results demonstrate that caspase-8 and -1 cleave pro–IL-1β at the same site.
Figure 4.
Poly(I:C)- and LPS-induced pro–IL-1β processing is caspase-8 dependent. (A) HEK293T cells were cotransfected with pro–IL-1β and either WT caspase-8 or caspase-8 C362A. 24 h later, processing of pro–IL-1β was analyzed by SDS-PAGE and Western blotting of total cell lysates (bottom). Secretion of biologically active IL-1β into the corresponding cell supernatants was analyzed via IL-1 bioassay (top). Expression of caspase-8 was verified by Western blotting. (B) Pro–IL-1β–transfected HEK293-TLR3 cells were incubated for 6 h with 25 μg/ml poly(I:C). 1 h before incubation, cells received 50 μM z-VAD-fmk, 50 μM z-IETD-fmk, or 0.05% DMSO (solvent control). Where indicated, cells were cotransfected with pCAGGS-CrmA-E (100 ng) or pCR3.1-Flag-vFLIP (50–100–200 ng). Pro–IL-1β processing and secretion of biologically active IL-1β was analyzed as described in A. (C) HEK293-TLR3 cells were transfected with either control nontargeting (N-i) or caspase-8 siRNA (C8-i). 72 h later, cells were treated for 6 h with 25 μg/ml poly(I:C) and analyzed for pro–IL-1β processing and secretion of biologically active IL-1β, as described in A. Knockdown of caspase-8 was verified by Western blotting. (D) RAW264.7 macrophages were transduced with either control nontargeting shRNA (Ctrl-sh) or caspase-8 shRNA (Casp-8-sh). 72 h later, cells were primed with BLP and subsequently incubated with poly(I:C), LPS, BLP, or control medium as described in the Materials and methods. Pro–IL-1β processing was analyzed by Western blotting of total cell lysates (TL) and IL-1β immunoprecipitates (IP) from the corresponding cell supernatant. Knockdown of caspase-8 was verified by Western blotting. Data are representative of two (C and D) or three (A and B) independent experiments.
Figure 5.
Recombinant caspase-8 cleaves pro–IL-1β in vitro. (A) [35S]methionine-labeled pro–IL-1β was incubated for 1.5 h at 37°C with increasing concentrations of recombinant caspase-8 (10, 30, 100, and 300 ng) or caspase-1 (30 ng). Pro–IL-1β cleavage was revealed by SDS-PAGE followed by autoradiography. (B) [35S]methionine-labeled pro–IL-1β WT or the indicated mutants were incubated for 1.5 h at 37°C with recombinant caspase-1, -3, or -8 (200 ng) and analyzed as in A.
In conclusion, the data described in this study demonstrate that TLR3 and TLR4 stimulation induces a TRIF-dependent signaling pathway that leads to the caspase-1–independent maturation of pro–IL-1β. More specifically, we provide evidence that caspase-8 mediates pro–IL-1β processing in response to TLR3 and TLR4 stimulation. Caspase-8 has been best characterized as a cysteine protease that cleaves specific substrates to transmit apoptotic signals downstream of death receptors. Additionally, roles for caspase-8 in mediating T, B and NK cell proliferation, macrophage differentiation, NF-κB activation, maintaining lymphocytes homeostasis, and suppressing immunodeficiency have become evident (3, 18). Full caspase-8 knockout mice are embryonic lethal, but tissue-specific inactivation has also revealed the key role of caspase-8 in innate and adaptive immunity (3, 18). The role described for caspase-8 in pro–IL-1β processing in macrophages further supports its immunoregulatory function. Because conditional deletion of caspase-8 in myeloid cells results in defective macrophage differentiation (18), we were unable to study pro–IL-1β processing in caspase-8 knockout macrophages. However, siRNA-mediated knockdown of caspase-8 completely prevented TLR3- and TLR4-induced pro–IL-1β processing in HEK293T cells, as well as in macrophages.
In contrast to our data that show a role for TRIF and caspase-8 in TLR3- and TLR4-induced pro–IL-1β processing, it has previously been reported by others that LPS and poly(I:C) can initiate caspase-1–mediated pro–IL-1β processing independent of TLRs or TLR-associated adaptor molecules (5, 19), but involving the NALP3 inflammasome (5, 8–10). It should be noted, however, that in the aforementioned reports, LPS- or poly(I:C)-induced caspase-1 activation required an additional pulse with ATP, which triggers a P2X7- and pannexin-1–mediated potassium efflux and cytosolic delivery of bacterial products to intracellular NALP3 (5). In fact, the essential role of NALP3 and caspase-1 in LPS-plus-ATP–induced pro–IL-1β processing was also confirmed in this study (Fig. 2 C and Fig. 3 A). In this context, it is worth mentioning that one of the original studies describing the caspase-1 knockout mice already reported that trace levels of mature IL-1β can be found in the supernatant of caspase-1–deficient macrophages stimulated for 30 min with LPS plus ATP, and further cultured for 3 h in the absence of ATP (20). Collectively, these data illustrate that differences in experimental conditions (e.g., costimulation with ATP or intracellular delivery of microbial products) might determine the signaling pathway that is used to induce pro–IL-1β processing.
Interestingly, caspase-1–independent pro–IL-1β processing has also been reported in turpentine- and zymosan-treated mice and macrophages (21), as well as in Fas-stimulated neutrophils (22). Consistent with the latter data, we were also able to show caspase-8 activation and pro–IL-1β processing in response to Fas overexpression in HEK293T cells (unpublished data). Because Fas-induced pro–IL-1β processing was previously reported to be sensitive to the pan-caspase inhibitor z-VAD.fmk, we speculate that caspase-8 is also involved in Fas-induced pro–IL-1β processing.
Consistent with the essential role of TRIF in TLR3- and TLR4-induced pro–IL-1β processing, we did not observe pro–IL-1β processing in response to TLR2, which does not signal via TRIF. Similarly, in contrast to TLR3 and TLR4, TLR2 stimulation did not induce caspase-8 processing, which is representative for caspase-8 activation (unpublished data). It is worth mentioning that caspase-8 has already been implicated in TLR3- and TLR4-induced signaling as part of a TRIF–RIP1-Fas-associated Death Domain-containing protein–caspase-8 complex leading to apoptosis (23, 24). In this context, we noticed that poly(I:C) and LPS also induced macrophage cell death upon longer incubation, especially when the synthesis of antiapoptotic proteins was inhibited by CHX (unpublished data). Although a detailed study of the underlying signaling mechanisms leading to caspase-8–mediated pro–IL-1β processing in response to TLR3 or TLR4 stimulation is beyond the scope of this study, it can be expected that a similar TRIF–RIP1–Fas-associated Death Domain-containing protein–caspase-8 pathway leads to pro–IL-1β processing. This is also supported by the specific coimmunoprecipitation of caspase-8 and its activated processing product with WT TRIF, but not with a RIP1 binding-deficient RHIM mutant of TRIF, from HEK293T cells (unpublished data). Collectively, these data implicate an important function for caspase-8 at the crossroads of proinflammatory and proapoptotic signaling. Classically, apoptosis is defined as a type of cell death that does not induce inflammation (25). This is most probably true in most cases of apoptosis, in which dying cells do not express pro–IL-1β or activated caspase-8. However, in pathological conditions associated with pro–IL-1β up-regulation and caspase-8 activation, inflammatory responses could be enhanced by the caspase-8 mediated processing and release of IL-1β from apoptotic macrophages. In view of the possible use of caspase-8 inhibitors as therapeutic agents, it will be important to identify those disease conditions in which caspase-8 is involved in the cleavage of pro–IL-1β.
MATERIALS AND METHODS
Cells, mice, antibodies, and reagents.
HEK293T, HEK293-TLR (provided by A. Chariot, University of Liege, Liege, Belgium), and RAW264.7 cells were grown in DMEM supplemented with 10% FCS, 2 mM l-glutamine, 0.4 mM sodium pyruvate, and antibiotics. Peritoneal macrophages were grown in RPMI 1640 supplemented with 1% FCS, 2 mM l-glutamine, 0.4 mM sodium pyruvate, antibiotics, and 50 μM β-mercaptoethanol. TRIF-deficient mice were obtained from B. Beutler (Scripps Research Institute, La Jolla, CA); caspase-1–deficient mice were obtained from R. Flavell (Yale University School of Medicine, New Haven, CT). TLR3-deficient mice were purchased from The Jackson Laboratory, and WT C57BL/6 mice were purchased from Janvier. Maintenance and care of mice complied with the guidelines of the University of Ghent Ethics Committee for the use of laboratory animals, which also approved the use of mice as a source of peritoneal macrophages. Other reagents used were poly(I:C) (GE Healthcare), Pam3Cys-SK4 (BLP; EMC Microcollections GmbH), ultrapure LPS (InvivoGen), CHX (Sigma–Aldrich), z-VAD-fmk (Bachem), Ac-WEHD-cho (Peptide Insitute, Inc.), z-DEVD-cmk (Bachem), and z-IETD-fmk (Merck). Recombinant murine caspase-1, -3, and -8 were provided by P. Vandenabeele (Ghent University, Ghent, Belgium). The antibodies used were as follows: anti-IL-1β (R&D Systems), mouse IL-1β antibody/IL-1F2 mAb (Clone 30311; R&D Systems), HRP-linked anti-E tag (GE Healthcare), HRP-linked anti-FLAG tag (Sigma-Aldrich), anti-HA tag (CRP), anti-actin (MP Biomedicals), anti–human caspase-8 (Invitrogen), anti–mouse caspase-8 (3B10; Qbiogene), HRP-linked anti–mouse Ig (GE Healthcare), HRP-linked anti–goat Ig (Santa Cruz Biotechnology, Inc.), HRP-linked goat anti–rat Ig (Southern Biotech).
Expression vectors.
pCR3.1-FLAG expression vectors for human TRIF, TRIF-TIR (aa 381–660), TRIF-NT (aa 1–385), TRIF-CT (aa 381–712), TRIF-RHIM (aa 534–712), and E8-vFLIP (Equine virus E8 protein) were a gift from J. Tschopp (University of Lausanne, Lausanne, Switzerland) (12). pcDNA3.1-HA-Mal was a gift from L. O'Neill (Trinity Colllege, Dublin, Ireland). Other used expression vectors are pCAGGS-E-CrmA (LMBP4888) and pCAGGS-pro–IL-1β (LMBP3853) (26), pCAGGS-E-MyD88 (27), pCDNA-mCASP-8 (LMBP3841), and pCAGGS-EmCASP-8-C362A (LMBP4942). pCAGGS-E-hTRIF (LMBP5226) and pCAGGS-E-hTRAM (LMBP5227) were prepared by cloning the human TRIF and TRAM cDNA, respectively, as a NotI–XbaI fragment into pCAGGS-E-tag. pGEM11-pro–IL-1β (LMBP4639) (28) encodes pro–IL-1β under control of an SP6 promotor. pCR3-pro–IL-1β-D105A, pCR3-pro–IL-1β-D108A, and pCR3-pro–IL-1β-D117A were generated by mutating murine pro–IL-1β cDNA from pGEM11-pro–IL-1β using overlap PCR, after which it was cloned into a pCR3 expression vector (Invitrogen) under the control of a T7 promotor. The plasmids that have been assigned a LMBP accession no. are available from the BCCM/LMBP Plasmid collection, Department of Molecular Biology, Ghent University, Belgium (http://bccm.belspo.be/about/lmbp.php).
Analysis of pro–IL-1β processing by Western blotting and immunoprecipitation.
HEK293T and HEK293-TLR3 cells (0.2–1 × 106/6-well) were cotransfected with pCAGGS-pro–IL-1β and other expression plasmids (1 μg DNA total) by calcium phosphate precipitation and Fugene (Roche), respectively. The next day, cells were stimulated and lysed at 4°C for 15 min in 100–200 μl lysis buffer (200 mM NaCl, 1% NP-40, 10 mM Tris-HCl, pH 7.5, 5 mM EDTA, and 2 mM DTT) supplemented with protease and phosphatase inhibitors. The cell lysates were subsequently separated by SDS-PAGE and analyzed by Western blotting and ECL detection (PerkinElmer). IL-1β was revealed with anti-IL-1β (R&D Systems). Biologically active IL-1 present in the cell supernatant was determined by bioassay, as previously described (29).
In the case of peritoneal macrophages, cells were pretreated for 8 h with 200 ng/ml BLP to up-regulate pro–IL-1β, and subsequently incubated for different times with 25 μg/ml poly(I:C), 100 ng/ml LPS, 200 ng/ml BLP, or control medium in the presence of 1 μg/ml CHX that was given 30 min before incubation. Endogenous pro–IL-1β was detected by Western blotting of total cell lysates. Mature IL-1β was detected in the cell supernatant via immunoprecipitation and Western blotting. Cell supernatants were incubated overnight with 10% lysis buffer and 2 μg mouse IL-1β antibody/IL-1F2 mAb (clone 30311; R&D Systems), followed by the incubation with protein G–Sepharose beads (GE Healthcare) for 3 h. The beads were washed four times with lysis buffer before elution with Laemmli buffer and further analysis by SDS-PAGE and Western blotting.
In vitro caspase cleavage assay.
pGEM11-pro–IL-1β, pCR3-pro–IL-1β-D105A, pCR3-pro–IL-1β-D108A, and pCR3-pro–IL-1β-D117A were used for in vitro–coupled transcription–translation of [35S]methionine-labeled pro–IL1-β in an in vitro reticulocyte lysate system (Promega) according to the manufacturer's protocol. Translation reactions (2 μl) were incubated with recombinant murine caspase-1, -3, or -8 in a total volume of 25 μl CFS buffer (10 mM Hepes-NaOH, pH 7.4, 220 mM mannitol, 68 mM sucrose, 2 mM NaCl, 2.5 mM KH2PO4, 0.5 mM EGTA, 2 mM MgCl2, 5 mM sodium pyruvate, 0.1 mM PMSF, and 1 mM DTT) for 1.5 h at 37°C. The resulting cleavage products were analyzed by SDS-PAGE and autoradiography.
RNA interference (RNAi).
HEK293-TLR3 cells were plated in 6-wells (2 × 105 cells/well) 72 h before transfection with human caspase-8 siRNA (ON-TARGETplus SMARTpoolhCASP8; Dharmacon) by DharmaFECT 1 (Dharmacon). 6 h after transfection, cells were split, and they were transfected via Fugene with pCAGGS-pro–IL-1β the following day. For RNAi in peritoneal macrophages, 106 cells were transfected with 2.5 μg nontargeting control (siCONTROL Non-Targeting siRNA Pool; Dharmacon), NALP3 siRNA (siRNA GenomeWide mouse NALP3; QIAGEN), or MDA5 siRNA (siRNA GenomeWide mouse MDA5; QIAGEN) using the Mouse Macrophage Nucleofector kit (Amaxa) according to the manufacturer's instructions. RNAi in RAW264.7 cells was obtained via lentiviral transduction. Lentivirus was produced by transfecting 2 × 106 HEK293T cells with 10 μg pLKO.1 control shRNA vector (Sigma-Aldrich; SHC002) or 10 μg pLKO.1 caspase-8 shRNA vector (Sigma-Aldrich; SHDNAC-TRCN0000012247), 3 μg pMD2-VSV, and 6.5 μg pCMVΔR8.91 (30) using the calcium-phosphate transfection method. 8 h after transfection, fresh medium was added to the cells, and 48 h later the viral supernatant was supplemented with 8 μg/ml polybrene, which was used to transduce 105 RAW264.7 cells in 6-wells.
RNA isolation, cDNA synthesis, and qPCR.
Total RNA was extracted from cells using the Aurum Total RNA Mini kit (Bio-Rad Laboratories) and reverse transcribed into cDNA with Superscript II Reverse transcription (Invitrogen) using oligo(dT)18 primers according to the manufacturer's instructions. qPCR was performed by using SYBR Green I Master mix (Roche) in the LightCycler 480 Detection System (Roche) with the following primers: HPRT, 5′-AGTGTTGGATACAGGCCAGAC-3′ and 5′-CGTGATTCAAATCCCTGAAGT-3′; NALP3, 5′-ATTACCCGCCCGAGAAAGG-3′ and 5′-TCGCAGCAAAGATCCACACAG-3′; MDA5, 5′-AGATCAACACCTGTGGTAACACC-3′ and 5′-CTCTAGGGCCTCCACGAACA-3′; IRF-7, 5′-CTGGAGCCATGGGTATGCA-3′ and 5′-AAGCACAAGCCGAGACTGCT-3′. Quantification was performed using the comparative CT method (ΔΔCT). Results are expressed relative to HPRT values.
Acknowledgments
A. Meeus, W. Burm, T. Boonefaes, and H. Revets are acknowledged for their technical assistance. We thank Dr. A. Chariot, Dr. L. O'Neill, Dr. J. Tschopp, Dr. B. Beutler, Dr. R. Flavell, and Dr. P. Vandenabeele for providing reagents.
This work was supported in part by grants from the Interuniversitaire Attractiepolen (IAP6/18), the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (FWO; grant 3G010505), and the Geconcerteerde Onderzoeksacties of the University of Ghent (GOA; grant 01G06B6). E. Vercammen and J. Maelfait are predoctoral research fellows with the FWO and the Instituut voor de aanmoediging van innovatie door Wetenschap en Technologie in Vlaanderen, respectively. S. Janssens and P. Schotte are postdoctoral research associates with the FWO.
The authors have no conflicting financial interests.
J. Maelfait and E. Vercammen contributed equally to this paper.
References
- 1.Dunne, A., and L.A. O'Neill. 2003. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci. STKE. 2003:re3. [DOI] [PubMed] [Google Scholar]
- 2.Martinon, F., and J. Tschopp. 2007. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 14:10–22. [DOI] [PubMed] [Google Scholar]
- 3.Lamkanfi, M., N. Festjens, W. Declercq, T. Vanden Berghe, and P. Vandenabeele. 2007. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 14:44–55. [DOI] [PubMed] [Google Scholar]
- 4.Creagh, E.M., and L.A. O'Neill. 2006. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 27:352–357. [DOI] [PubMed] [Google Scholar]
- 5.Kanneganti, T.D., M. Lamkanfi, Y.G. Kim, G. Chen, J.H. Park, L. Franchi, P. Vandenabeele, and G. Nunez. 2007. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity. 26:433–443. [DOI] [PubMed] [Google Scholar]
- 6.Martinon, F., L. Agostini, E. Meylan, and J. Tschopp. 2004. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol. 14:1929–1934. [DOI] [PubMed] [Google Scholar]
- 7.Pan, Q., J. Mathison, C. Fearns, V.V. Kravchenko, J.D. Correia, H.M. Hoffman, K.S. Kobayashi, J. Bertin, E.P. Grant, A.J. Coyle, et al. 2007. MDP-induced interleukin-1β processing requires Nod2 and CIAS1/NALP3. J. Leukoc. Biol. 82:177–183. [DOI] [PubMed] [Google Scholar]
- 8.Kanneganti, T.D., M. Body-Malapel, A. Amer, J.H. Park, J. Whitfield, L. Franchi, Z.F. Taraporewala, D. Miller, J.T. Patton, N. Inohara, and G. Núñez. 2006. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281:36560–36568. [DOI] [PubMed] [Google Scholar]
- 9.Mariathasan, S., D.S. Weiss, K. Newton, J. McBride, K. O'Rourke, M. Roose-Girma, W.P. Lee, Y. Weinrauch, D.M. Monack, and V.M. Dixit. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 440:228–232. [DOI] [PubMed] [Google Scholar]
- 10.Sutterwala, F.S., Y. Ogura, M. Szczepanik, M. Lara-Tejero, G.S. Lichtenberger, E.P. Grant, J. Bertin, A.J. Coyle, J.E. Galán, P.W. Askenase, and R.A. Flavell. 2006. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 24:317–327. [DOI] [PubMed] [Google Scholar]
- 11.Kawai, T., and S. Akira. 2007. TLR signaling. Semin. Immunol. 19:24–32. [DOI] [PubMed] [Google Scholar]
- 12.Meylan, E., K. Burns, K. Hofmann, V. Blancheteau, F. Martinon, M. Kelliher, and J. Tschopp. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-κB activation. Nat. Immunol. 5:503–507. [DOI] [PubMed] [Google Scholar]
- 13.Sato, S., M. Sugiyama, M. Yamamoto, Y. Watanabe, T. Kawai, K. Takeda, and S. Akira. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304–4310. [DOI] [PubMed] [Google Scholar]
- 14.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K.J. Ishii, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105. [DOI] [PubMed] [Google Scholar]
- 15.Callus, B.A., and D.L. Vaux. 2007. Caspase inhibitors: viral, cellular and chemical. Cell Death Differ. 14:73–78. [DOI] [PubMed] [Google Scholar]
- 16.Hazuda, D.J., J. Strickler, F. Kueppers, P.L. Simon, and P.R. Young. 1990. Processing of precursor interleukin 1β and inflammatory disease. J. Biol. Chem. 265:6318–6322. [PubMed] [Google Scholar]
- 17.Howard, A.D., M.J. Kostura, N. Thornberry, G.J. Ding, G. Limjuco, J. Weidner, J.P. Salley, K.A. Hogquist, D.D. Chaplin, R.A. Mumford, et al. 1991. IL-1-converting enzyme requires aspartic acid residues for processing of the IL-1β precursor at two distinct sites and does not cleave 31-kDa IL-1α. J. Immunol. 147:2964–2969. [PubMed] [Google Scholar]
- 18.Kang, T.B., T. Ben-Moshe, E.E. Varfolomeev, Y. Pewzner-Jung, N. Yogev, A. Jurewicz, A. Waisman, O. Brenner, R. Haffner, E. Gustafsson, et al. 2004. Caspase-8 serves both apoptotic and nonapoptotic roles. J. Immunol. 173:2976–2984. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto, M., K. Yaginuma, H. Tsutsui, J. Sagara, X. Guan, E. Seki, K. Yasuda, M. Yamamoto, S. Akira, K. Nakanishi, et al. 2004. ASC is essential for LPS-induced activation of procaspase-1 independently of TLR-associated signal adaptor molecules. Genes Cells. 9:1055–1067. [DOI] [PubMed] [Google Scholar]
- 20.Li, P., H. Allen, S. Banerjee, S. Franklin, L. Herzog, C. Johnston, J. McDowell, M. Paskind, L. Rodman, J. Salfeld, et al. 1995. Mice deficient in IL-1β-converting enzyme are defective in production of mature IL-1β and resistant to endotoxic shock. Cell. 80:401–411. [DOI] [PubMed] [Google Scholar]
- 21.Fantuzzi, G., G. Ku, M.W. Harding, D.J. Livingston, J.D. Sipe, K. Kuida, R.A. Flavell, and C.A. Dinarello. 1997. Response to local inflammation of IL-1β-converting enzyme- deficient mice. J. Immunol. 158:1818–1824. [PubMed] [Google Scholar]
- 22.Miwa, K., M. Asano, R. Horai, Y. Iwakura, S. Nagata, and T. Suda. 1998. Caspase 1-independent IL-1β release and inflammation induced by the apoptosis inducer Fas ligand. Nat. Med. 4:1287–1292. [DOI] [PubMed] [Google Scholar]
- 23.Han, K.J., X. Su, L.G. Xu, L.H. Bin, J. Zhang, and H.B. Shu. 2004. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-κB activation and apoptosis pathways. J. Biol. Chem. 279:15652–15661. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser, W.J., and M.K. Offermann. 2005. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J. Immunol. 174:4942–4952. [DOI] [PubMed] [Google Scholar]
- 25.Henson, P.M., D.L. Bratton, and V.A. Fadok. 2001. Apoptotic cell removal. Curr. Biol. 11:R795–R805. [DOI] [PubMed] [Google Scholar]
- 26.Schotte, P., G. Denecker, A. Van Den Broeke, P. Vandenabeele, G.R. Cornelis, and R. Beyaert. 2004. Targeting Rac1 by the Yersinia effector protein YopE inhibits caspase-1-mediated maturation and release of interleukin-1β. J. Biol. Chem. 279:25134–25142. [DOI] [PubMed] [Google Scholar]
- 27.Janssens, S., K. Burns, J. Tschopp, and R. Beyaert. 2002. Regulation of interleukin-1- and lipopolysaccharide-induced NF-κB activation by alternative splicing of MyD88. Curr. Biol. 12:467–471. [DOI] [PubMed] [Google Scholar]
- 28.Beyaert, R., V.J. Kidd, S. Cornelis, M. Van de Craen, G. Denecker, J.M. Lahti, R. Gururajan, P. Vandenabeele, and W. Fiers. 1997. Cleavage of PITSLRE kinases by ICE/CASP-1 and CPP32/CASP-3 during apoptosis induced by tumor necrosis factor. J. Biol. Chem. 272:11694–11697. [DOI] [PubMed] [Google Scholar]
- 29.Vandenabeele, P., W. Declercq, C. Libert, and W. Fiers. 1990. Development of a simple, sensitive and specific bioassay for interleukin-1 based on the proliferation of RPMI 1788 cells. Comparison with other bioassays for IL-1. J. Immunol. Methods. 135:25–32. [DOI] [PubMed] [Google Scholar]
- 30.Zufferey, R., D. Nagy, R.J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871–875. [DOI] [PubMed] [Google Scholar]