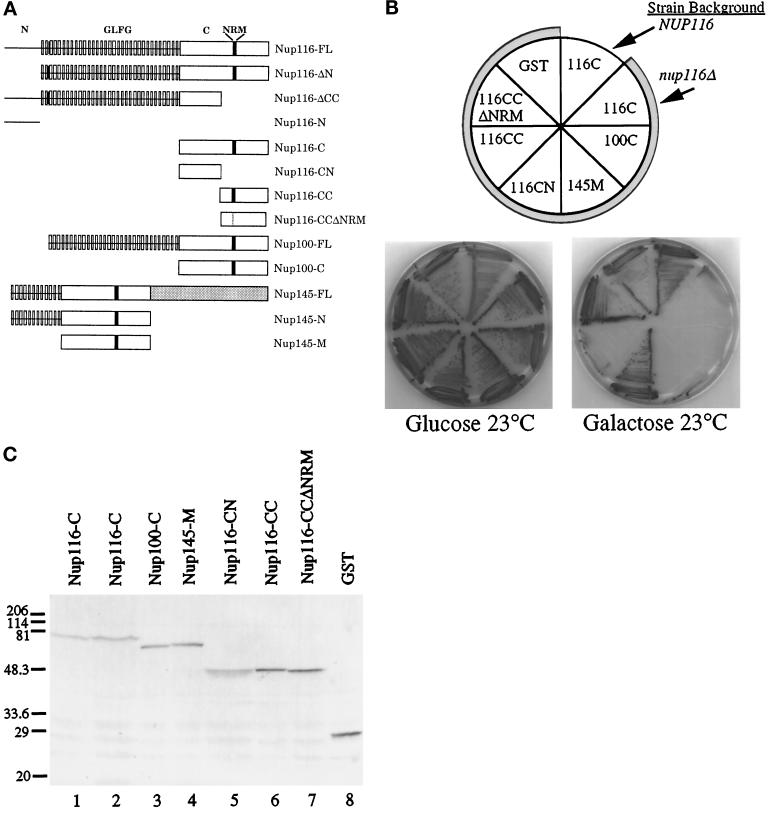
Figure 1.
The nup116-C lethal phenotype. (A) Schematic diagrams of the structural regions of Nup116p, Nup100p, and Nup145p and their related deletion/truncation polypeptides as referred to in this study. The N-terminal region of Nup116p spans amino acid residues 1–180, GLFG region residues 181–725, the C-terminal region residues 726-1113, the CN region residues 726–919, and the CC region residues 914-1113. The similarity between the C-terminal regions of Nup116p and Nup100p is 78%, whereas among Nup116-C, Nup100-C, and the middle region of Nup145 (Nup145-M) it is 55% (Wente et al., 1992; Fabre et al., 1994; Wente and Blobel, 1994). The NRM region in Nup116p spans residues 998-1005 (Fabre et al., 1994). FL, full- length protein. (B) The nup116Δ strain is inviable when the homologous Nup116-C, Nup100-C, and Nup145-M regions fused to GST are expressed from a galactose-inducible promoter. The plasmids expressing the indicated protein regions (see Table 1) were transformed into W303α (NUP116) or SWY27 (nup116Δ::HIS3) (shaded sectors). Strains were grown at 23°C for 7 d on SM-trp, 2% glucose, or 2% galactose. Expression of Nup116-C, Nup100-C, and Nup145-M inhibited colony formation of nup116Δ cells. Expression of constructs with a deletion of the NRM (Nup116-CCΔNRM) in the carboxyl-terminal half of Nup116-C or with only the amino-terminal half (Nup116-CN) present resulted in viability. Expression of GST alone did not inhibit growth of the nup116Δ strain. (C) Immunoblot analysis of the strains shown in B. Lane 1, Nup116-C expression in NUP116 cells (W303α). Lanes 2–8, expression in nup116Δ cells (SWY27). Yeast lysates were separated on a 10.5% SDS-polyacrylamide gel and transferred to nitrocellulose for immunoblotting with an affinity-purified rabbit polyclonal antibody recognizing GST. Molecular mass markers are in kDa.