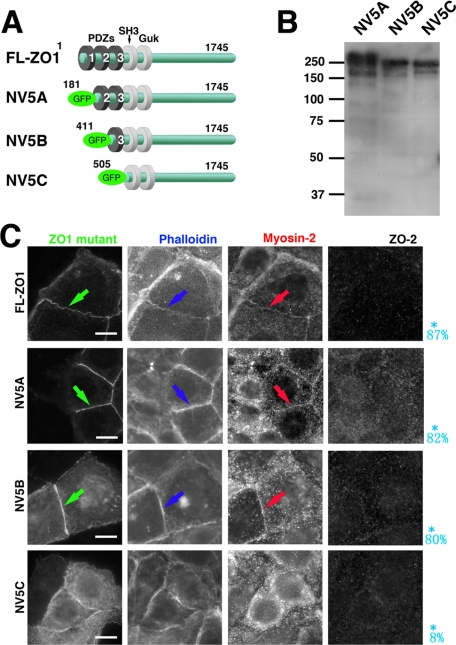
Figure 4.
Role of PDZ-3 domain of ZO-1 in restoration of formation of ZA. (A) Schematic drawing which shows constructs of ZO-1 deletion mutants. ZO-1 deletion mutants 5A (181-1745 a.a.), 5B (411-1745 a.a.), and 5C (505-1745 a.a.) were generated by PCR amplification and subcloned into CAG-N-Venus vector, producing GFP-NV5A(NV5A), GFP-NV5B(NV5B), and GFP-NV5C(NV5C) constructs, respectively. (B) Immunoblotting of total cell lysates of ZO1(ko)/2(kd) Eph4 cells transiently expressing each mutant of NV5A, NV5B, and NV5C. Detection by anti-GFP mAb. (C) Immunofluorescence of ZO1(ko)/2(kd) Eph4 cells transiently expressing each mutant of NV5A, NV5B, and NV5C for myosin-2, ZO-2, and fluorescence for GFP-ZO-1 mutant and actin due to phalloidin-staining. The mutants of ZO-1 lacking the PDZ-1 or PDZ-1/2 domains (NV5A and NV5B) are localized in ZA positive for myosin-2, similarly effective for junctional integration of myosin-2 into ZA (arrows). In contrast, the mutant lacking the PDZ1/2/3 domain of (NV5C), though localized in prezonula-ZA, does not restore junctional integration of myosin-2, resulting in combination of myosin-2–negative prezonula-AJ and cytoplasmic actomyosin-positive ring like structures. The probability of integration of myosin-2 into cell–cell AJ to form ZA, reduced from 20 transfectants in the immunofluorescence micrographs, are shown in blue letters. Bars, 5 μm.