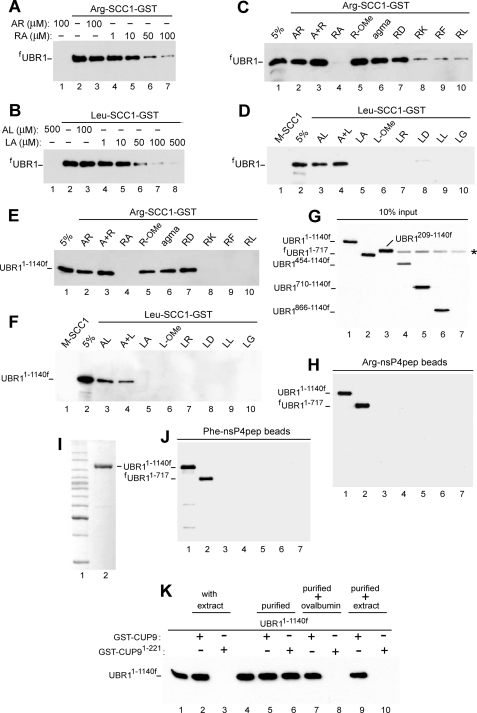
FIGURE 4.
Specific interactions between UBR1 and its physiological substrates or model ligands. A, GST pulldown assays with full-length, FLAG-taggedfUBR1 and immobilized Arg-SCC1-GST. Equal amounts of extract from S. cerevisiae that expressed fUBR1 were incubated with glutathione-Sepharose beads preloaded with Arg-SCC1-GST, in the presence of indicated concentrations of either the Arg-Ala (RA) or Ala-Arg (AR) dipeptides. The beads-associated fUBR1 was eluted from the beads, fractionated by SDS-PAGE, and detected by immunoblotting with anti-FLAG antibody (see “Experimental Procedures”). B, same as in A but with Leu-SCC1-GST and either Leu-Ala (LA) or Ala-Leu (AL). C, same assay format, with Arg-SCC1-GST. Lane 1, 5% refers to a directly loaded sample of fUBR1-containing yeast extract that corresponded to 5% of the amount of extract in GST pulldowns of this panel. Lane 2, Arg-SCC1-GST pulldown assay with fUBR1 in the presence of 1 mm AR dipeptide (single-letter notations for amino acids). Lanes 3-10, same as lane 2 but with either A+R(1 mm free Ala and 1 mmfree Arg), or RA, or R-OMe (O-methyl ester), or agmatine (see the main text), or RD, or RK, or RF, or RL, all of them at 1 mm. D, same as in C but with Leu-SCC1-GST and different dipeptides or related compounds, as shown. Lane 1, Met-SCC1-GST (instead of Leu-SCC1-GST); note the absence of its interaction with fUBR1, in contrast to Leu-SCC1-GST and Arg-SCC1-GST. E, same as in C, but with UBR11-1140f, the N-terminal half of UBR1. F, same as in D but with UBR11-1140f. G, input samples of UBR11-1140f and its derivatives. Lane 1, UBR11-1140f. Lanes 2-7, fUBR11-717, UBR1209-1140f, UBR1454-1140f, UBR1710-1140f, UBR1866-1140f, and UBR1ha, respectively. These derivatives of UBR1 were overexpressed in S. cerevisiae as described for UBR11-1140f. The asterisk denotes a protein cross-reacting with anti-FLAG antibody. UBR1ha (lane 7) was employed as a (negative) control for antibody specificity. 10% input refers to a directly loaded sample of yeast extract that corresponded to 10% of the amount of extract in GST pulldowns of H and J. H, X-peptide assay (see “Experimental Procedures”) with UBR11-1140f and its derivatives, described in G. UBR11-1140f and fUBR11-717, but not the other UBR1 derivatives of G bound to immobilized Arg-nsP4pep peptide. I, lane 1, molecular mass markers. Lane 2, Coomassie-stained SDS-PAGE pattern of (overexpressed) UBR11-1140f that was purified from yeast extract using anti-FLAG affinity chromatography and used for GST-CUP9 pulldowns (see K and “Experimental Procedures”). J, same as in H but with immobilized Phe-nsP4pep peptide. K, lane 1, 2% input. Lane 2, GST-CUP9 pulldown with UBR11-1140f in S. cerevisiae extract. Lane 3, same as lane 2, but with GST-CUP91-221, a C-terminally truncated mutant of the 306-residue CUP9 that lacks its C-terminal degron (67). Note the absence of binding of UBR11-1140f (in yeast extract) to GST-CUP91-221, in contrast to its binding to full-length GST-CUP91-306. Lane 4, 10% input. Lane 5, same as lane 2 but with purified UBR11-1140f, as distinguished from UBR11-1140f in yeast extract. Lane 6, same as lane 5 but with GST-CUP91-221. Note that purified UBR11-1140f, in contrast to the same protein in yeast extract, binds to both degron-containing (lane 5) and degron-lacking CUP9 (lane 6). Lane 7, same as lane 5 but with ovalbumin (at 50 mg/ml) added to UBR11-1140f before GST pulldown with full-length GST-CUP9. Lane 8, same as lane 7 but with GST-CUP91-221. Note restoration of the binding specificity of UBR11-1140f in the presence of ovalbumin. Lanes 9 and 10, same as lanes 5 and 6 but “empty” yeast extract was added to purified UBR11-1140f before GST pulldowns with either full-length GST-CUP9 (lane 9) or degron-lacking GST-CUP91-221.