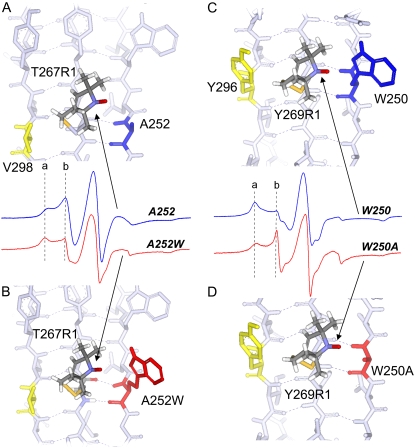
FIGURE 6.
(A) Structure and EPR spectrum (blue) of T267R1 with wild-type HB and nonhydrogen-bonded neighbors A252 and V298 (in blue and yellow), respectively. (B) Structure and EPR spectrum (red) of T267R1 with HB neighbor mutated to tryptophan (A252W, in red). (C) Structure and EPR spectrum (blue) of T269R1 with wild-type HB and nonhydrogen-bonded neighbors W250 and Y296 (blue and yellow), respectively. (D) Structure and EPR spectrum (red) of T269R1 with HB neighbor mutated to alanine (W250A in red). Structures are based on the apo BtuB crystal structure (Protein Data Bank identification, 1NQE), with the spin-labeled side chain, R1, substituted as shown. The first R1 side-chain dihedral angles, χ1 and χ2, were set in the {m,m} rotameric state (37). Dashed vertical lines a and b result from immobile and mobile motional modes in the R1 side chain, respectively.