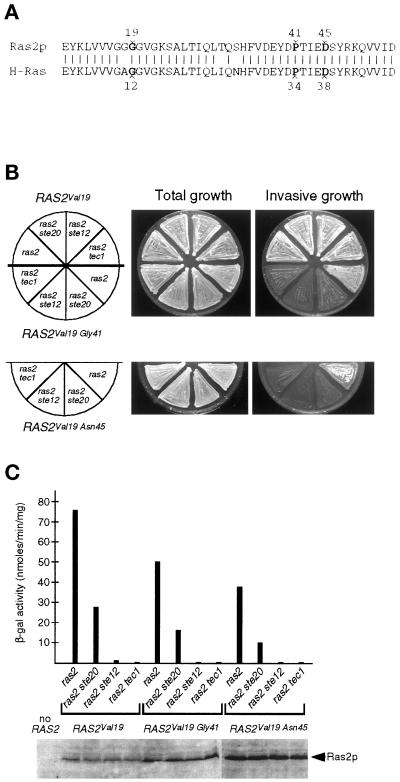
Figure 5.
Properties of RAS2 mutants. (A) Sequence alignment between N-terminal regions of yeast Ras2p (amino acids 10–75) and human H-Ras (amino acids 3–68). Identical residues are indicated by vertical lines. Substitution of Gly19 in yeast Ras2p or Gly12 in H-Ras for Val leads to activated mutant proteins. Amino acids Pro34 and Asp38 of H-Ras and corresponding residues Pro41 and Asp45 of Ras2p are indicated. (B) Invasive growth of yeast strains YHUM120 (ras2), YHUM158 (ras2 ste20), YHUM175 (ras2 ste12), and YHUM167 (ras2 tec1) containing the plasmid YCplac22-RAS2Val19 (RAS2Val19), YCplac22-RAS2Val19Gly41 (RAS2Val19Gly41) or YCplac22-RAS2Val19Asn45 (RAS2Val19Asn45) on solid SC-Trp medium. After incubation, plates were photographed before (Total growth) and after (Invasive growth) washing under a stream of water. (C) Expression levels of FRE(Ty1)::lacZ and Ras2 proteins. β-Galactosidase activity was measured on strains described in B, and expression of Ras2 mutant proteins was analyzed by Western blot analysis.