Fig. 1.
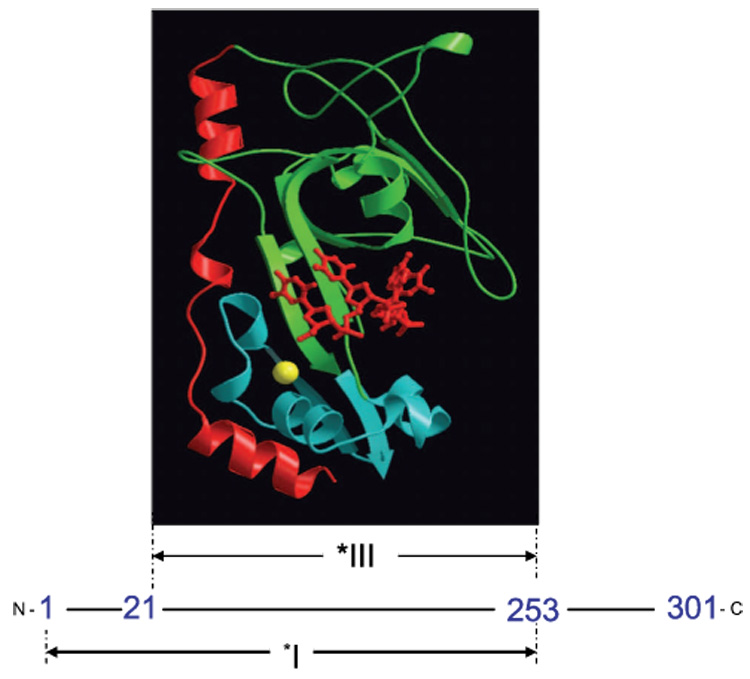
Proteolytic fragments of gene 32 protein. *I is obtained by trypsin cleavage of full length gp32 at residue 253, while *III results from cleavage at residues 21 and 253. A MOLSCRIPT 63 representation of a *III-oligonucleotide complex is shown at its location within the protein sequence. The protein is pictured in ribbon mode, with the major lobe green, the minor (Zn-containing) lobe blue, and the residue 198–239 flap red. The bound oligonucleotide, in sticks mode, is red, and the coordinated Zn2+, in space-filling mode, is yellow. The position of the oligodeoxynucleotide, pTTAT, is approximate; it was modeled by Shamoo et al. to maximally overlap excess electron density in the trough 64. The Protein Data Bank entry for core domain (without the oligonucleotide) is 1gpc.pdb.
