Figure 8.
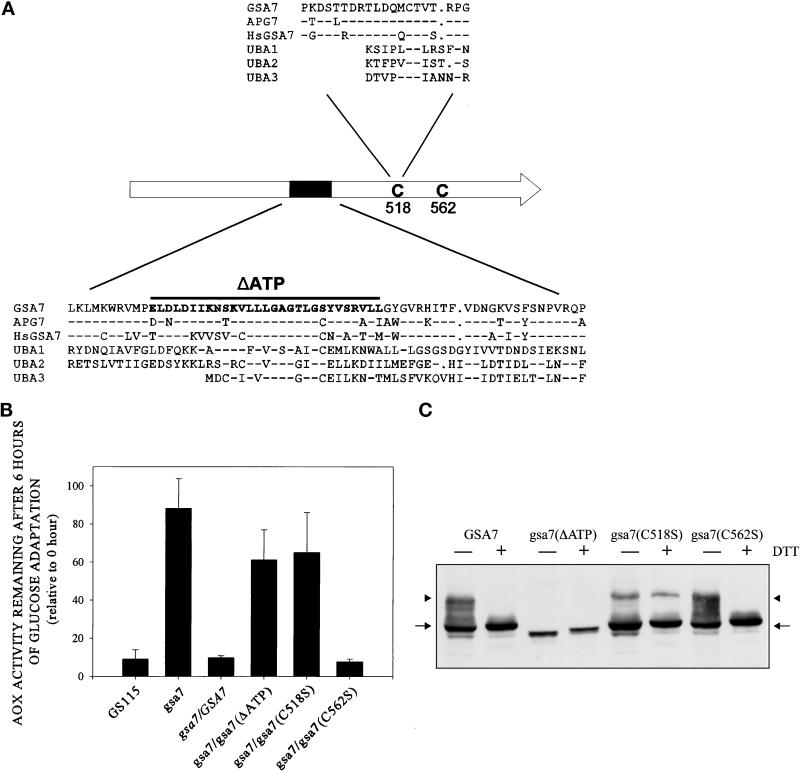
Complementation of gsa7 by normal and mutant forms of recombinant Gsa7-HA. (A) GSA7, APG7, and HsGSA7 proteins were aligned to three ubiquitin-activating enzymes (UBA1, UBA2, and UBA3). The amino acid alignment around the putative catalytic domain (C518) and ATP-binding region (K327–R342) are indicated. Amino acid identity represented by dashed lines was evident within these regions. Gaps represented by dots were inserted to optimize amino acid alignments. (B) gsa7 cells were stably transformed with GSA7-HA, gsa7(ΔATP), gsa7(C518S), and gsa7(C562S). The cells were then grown in methanol induction medium then adapted to glucose for 6 h. Cell extracts were prepared, and AOX assays were performed. The resulting values are presented as a percentage of the activity measured at 0 h and represent the mean ± SD of four or more determinations. (C) Aliquots of cell extracts prepared at 3 h of glucose adaptation were solubilized in 2% SDS and boiled for 3 min (−DTT) or solubilized in 2% SDS with 1.5% DTT and boiled for 5 min (+DTT), and the proteins were separated by SDS-PAGE. After transfer to nitrocellulose, the epitope-tagged Gsa7 proteins were identified using polyclonal antibodies that recognized the HA epitope. Gsa7-HA migrated as a 72-kDa protein (arrow). A thio-ester conjugate of Gsa7-HA was observed at ∼100 kDa (arrowhead).