Figure 4.
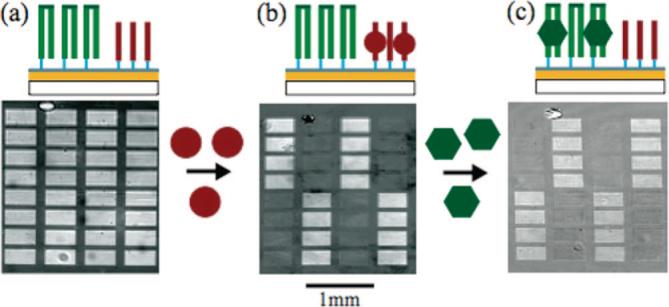
Protein-DNA binding experiment. (a) SPRi image of an oligonucleotide array consisting of two oligonucleotide sequences photolithographically synthesized on an amorphous carbon substrate. The double-stranded oligonucleotide sequences are hairpin structures (green) formed by synthesizing a self-complementary oligonucleotide sequence containing a TCCT hairpin loop sequence. The second oligonucleotide sequence (red) had no self- and/or interstrand complementary sequences, such that it remained single-stranded throughout the experiment. Hairpin structures were formed by incubating the surface at 90 °C followed by a slow cooling process. (b) First, the surface was exposed to a 10.0 μg/mL thrombin solution and binding to the single-stranded target oligonucleotide sequence was monitored with SPRi. The difference image shown was obtained by subtracting the image after thrombin binding from the reference image taken prior to binding. (c) Next a 1.0 μg/mL solution of the VFR protein was exposed to the oligonucleotide array and binding was monitored. The difference image was obtained by subtracting the image after VFR binding from the image after thrombin binding.
