Fig. 3.
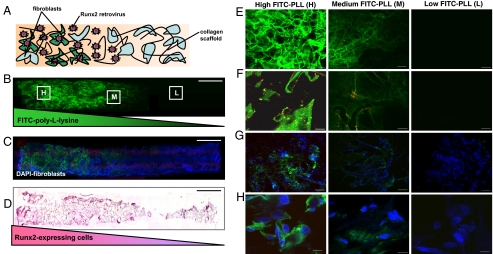
Spatially regulated genetic modification of fibroblasts within 3D matrices. (A) Schematic representation of a fibroblast-seeded construct containing spatial patterns of noncovalently immobilized retrovirus. A spatial distribution of Runx2 retrovirus (R2RV) was created by partially coating the proximal portion (left side) of collagen scaffolds with PLL at a dipping speed of 170 μm/s before incubation in retroviral supernatant and cell seeding. (B and C) Confocal microscopy images demonstrating a graded distribution of FITC-labeled PLL (green) (B) and FITC-labeled PLL gradient colocalized with uniformly distributed cell nuclei (DAPI, blue) (C). (Scale bar: 2 mm.) (D) Immunohistochemical staining for eGFP (pink) counterstained with hematoxylin (blue) revealed elevated eGFP expression on the proximal scaffold portion coated with PLL+R2RV. (Scale bar: 2 mm.) (E–H) High magnification confocal microscopy images moving lengthwise down the z-axis of construct shown in (B) at high (H), medium (M), and low (L) PLL densities. (E) FITC-labeled-PLL density is controlled by incubation/residence time in coating solution according to dipping speed. (Scale bar: 100 μm.) (F) Red fluorescent protein-labeled retrovirus (RFP-RV) colocalizes with FITC-PLL, confirming PLL-mediated immobilization of retrovirus. (Scale bars: 10 μm.) (G and H) Spatial control over retroviral transduction was assessed by coating scaffolds with a gradient of unlabeled PLL and unlabeled R2RV before cell seeding. High levels of eGFP expression (green) in DAPI-stained cells (blue) indicated efficient retroviral transduction in construct region containing high PLL density (H). (Scale bars: G, 100 μm; H, 10 μm.)