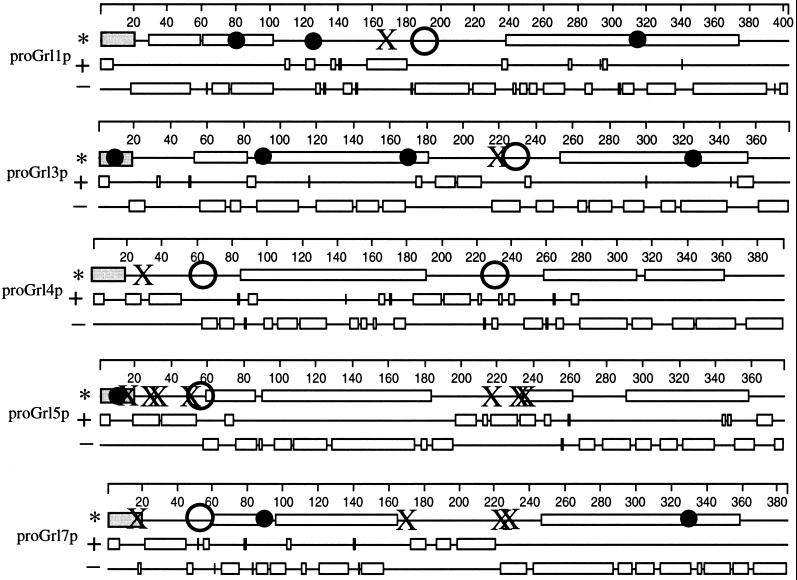
Figure 3.
Predicted features of proGrlps. On the basis of the deduced amino acid sequences and the known N-termini of isolated polypeptides, a number of features are predicted. Features of each Grlp are shown on three consecutive lines. On the first of each set of three lines are shown the regions forming coiled-coils (as open rectangles), sites corresponding to known amino termini of processed polypeptides (as large open circles), cysteine residues (as small solid circles), methionines (as X’s), and signal sequences (as shaded rectangles). On the second line, stretches of basic residues are shown as open rectangles. On the third line, stretches of acidic residues are shown as open rectangles.