Abstract
Inferring evolutionary relationships among highly divergent protein sequences is a daunting task. In particular, when pairwise sequence alignments between protein sequences fall <25% identity, the phylogenetic relationships among sequences cannot be estimated with statistical certainty. Here, we show that phylogenetic profiles generated with the Gestalt Domain Detection Algorithm–Basic Local Alignment Tool (GDDA-BLAST) are capable of deriving, ab initio, phylogenetic relationships for highly divergent proteins in a quantifiable and robust manner. Notably, the results from our computational case study of the highly divergent family of retroelements accord with previous estimates of their evolutionary relationships. Taken together, these data demonstrate that GDDA-BLAST provides an independent and powerful measure of evolutionary relationships that does not rely on potentially subjective sequence alignment. We demonstrate that evolutionary relationships can be measured with phylogenetic profiles, and therefore propose that these measurements can provide key insights into relationships among distantly related and/or rapidly evolving proteins.
Keywords: ab initio, retroelements, reverse transcriptase, GDDA-BLAST
The “protein problem” has remained unsolved despite decades of research (1, 2). In principle, one expects that the primary amino acid sequence of a protein determines its structure, function, and evolutionary (SF&E) characteristics. Yet, there still is no reliable method for predicting the native state structure of a protein and its function given only its sequence. In addition, inferring the evolutionary relationships among highly divergent protein and/or rapidly evolving sequences is a daunting task. In general, when pairwise sequence alignments between protein sequences fall below ≈25% identity (i.e., the “twilight zone”), the assignment of positional homology is so difficult that it becomes impossible to safely estimate phylogenetic relationships (1, 3, 4). However, a small number of conserved residues (≈8% identity) can coordinate the 3-D fold and/or function of proteins (5–7). Conversely, two proteins that share 88% identity can still retain independent structure and function (8).
The aforementioned studies point out that quantitatively measuring data spaces in the protein world (i.e., the sequence, structure, and functional space that proteins occupy) is a fundamental question facing evolutionary/computational biologists, with further questions arising. Is there any equation that quantitatively connects these protein spaces to protein evolution? Which residues within amino acid sequences best reflect the evolutionary history of a given protein? Do proteins with similar sequence and structure necessarily share a common ancestor? Furthermore, if sequence and structure similarity suggest an evolutionary history, can weak similarities be strengthened by functional connections? All of these questions are essentially connected to the protein data space; however, to date they have not been clearly solved either experimentally or theoretically.
Common computer alignment programs such as BLAST and FASTA (9, 10) often fail to detect distant protein relationships with sufficient statistical significance (3). This has spurred a Herculean effort from many researchers to develop strategies for detection of statistically significant similarities between distantly related proteins. For example, Blake and Cohen (11) built new amino acid substitution matrices for improving the accuracy of sequence alignment. More recently, advanced sequence comparison methods have been developed on the basis of shared features of sets of related sequences such as protein families. Examples of such approaches are templates (12, 13), profiles (14–16), hidden Markov models [HMMs; (17, 18)], and PSI-BLAST [position-specific iterated BLAST (19)]. In addition, threading algorithms are also intended to improve detection of homologous pairs from the sequence space in the twilight zone. Most of them were reported to show higher quality detection of evolutionary relationships of proteins compared with previous methods. In particular, Park et al. (3) reported that the SAM-T98 and PSI-BLAST (sequence profile-based algorithm) show threefold higher performance to detect the homologous relationships—between the sequences with <30% sequence identity—compared with the ordinary pairwise methods of FASTA and gapped BLAST (19). They determined that comparison of query sequences with models of clustered multiple sequence information can be more sensitive than using a single sequence alone. This is also true of LEK clustering, which performs and all-against-all comparison of sequences within a dataset (20), and has been successfully used to determine divergent evolutionary relationships (21).
What unites all of these methods is their exploitation of information encoded in multiple sequences. The recent explosion in the availability of knowledge bases and computational techniques for the analysis of complex data has created an unprecedented opportunity for teasing out invaluable information from protein sequences. Starting with a basic premise that protein sequence encodes information about SF&E, we proposed that phylogenetic profiles provide a unified framework for inferring SF&E from sequence information (22). Herein we demonstrate that the gestalt domain detection algorithm–basic local alignment tool (GDDA-BLAST) generates phylogenetic profiles that have the capacity to derive phylogenetic relationships for highly divergent proteins in a quantifiable manner, entirely independent of multiple sequence alignment (MSA). Results from our computational case study on a benchmark set of the highly divergent family of retroelements generally accord with those previously reported, and demonstrate that GDDA-BLAST measurements can be treated as “fingerprints” that can be used to derive distance estimates, and thus phylogenetic relationships, without prior information.
Results
Retroelements Are a Benchmark Set for the Twilight Zone of Sequence Similarity.
Self-replicating genetic elements such as retrotransposons use reverse transcriptase (RT, an RNA-dependent DNA polymerase) to multiply via an RNA intermediate copied into DNA (23). These highly diverse and likely ancient proteins are extremely effective to replicate and, along with other transposable elements, make up ≈50% of eukaryotic genomes by weight (23, 24). The first RT was discovered as a retroviral encoded enzyme (25). Subsequently, multiple genetic elements from diverse organisms have been shown to encode proteins that share sequence similarity to the retroviral RT, including cellular telomerase (23). Given (a) the >20 years of research/literature on this protein family, (b) the extreme nature of divergence within the known family members, and (c) its implications for the early evolution of life on earth and major infectious diseases of humans (26), retroelements are an excellent and rigorous benchmark set to test whether phylogenetic profiles can measure evolutionary distances.
The RT domain is the only known region in common to all classes of retroelements and therefore is often used for comparative analysis (23, 27, 28). Within the highly divergent RT domain, seven conserved motifs in the catalytic region of the enzyme have been identified that enable phylogenetic inferences of retrotransposons (23, 27, 29, 30). However, limiting the alignment space to these motifs requires potentially subjective manual editing, generating few phylogenetically informative sites. Thus, deep phylogenetic relationships are often ambiguous at best. Indeed, even these seven conserved motif are as divergent as their functional constraints will allow (23, 31). As a consequence, the precise phylogenetic relationship of the retroelements is still a subject of debate.
We curated 88 RT-containing sequences representing 11 groups of retroelements [see supporting information (SI) Table S1 for complete description and ref. 23 for excellent review]. The individual groups are from a broad range of taxa and comprise (i) long-terminal-repeat RTs (LTR: containing Ty1/Copia, Ty3/Gypsy, and BEL/Pao subgroups); (ii) retroviruses (e.g., HIV); (iii) pararetroviruses (RT-containing DNA viruses), including hepadnaviruses of animals (e.g., hepatitis B) and caulimoviruses of plants (e.g., cauliflower mosaic virus), (iv) tyrosine recombinase RTs (e.g., DIRS-1 elements); (v) non-LTR retrotransposons; (vi) Penelope-like elements (PLE); (vii) telomerases (TERT); (viii) group II introns (i.e., retrointrons); (ix) Mt Plasmids (i.e., retroplasmids); (x) Ms DNAs (i.e., retrons); and (xi) diversity-generating retroelements (DGR) (23, 24, 32–34).
Quantitative analysis of within-group and between-group variations in sequence similarity was performed by using global pairwise alignments on the RT domain (see Materials and Methods, Table S2, and Fig. S1). Overall, the average percentage identity between 88 RT sequences was 17.7% (± 6.0% s.d.). Specifically, 3,644 pairs (95.2%) among 3,828 possible pairs of these 88 sequences have <25% sequence identity. Within our dataset, the group with the highest sequence identity is the Mt plasmid group (mean 61.2% identity ± 41.8% s.d.), and the group with the smallest sequence identity is the telomerase group (mean 17.9% identity ± 4.4% s.d.). As a whole, RT sequences reside in the twilight zone of sequence similarity, underscoring the reason why deducing evolutionary relationships within the RT family is extremely challenging. We sought a method independent of multiple sequence alignment for inferring homology in this key protein family, based on our central hypothesis that profile-sequence alignments below statistical thresholds are not necessarily random, can retain biologically relevant information that is not reflected by chance similarity, and can be encoded into an informative phylogenetic profile matrix.
Phylogenetic Profiles and GDDA-BLAST Concept.
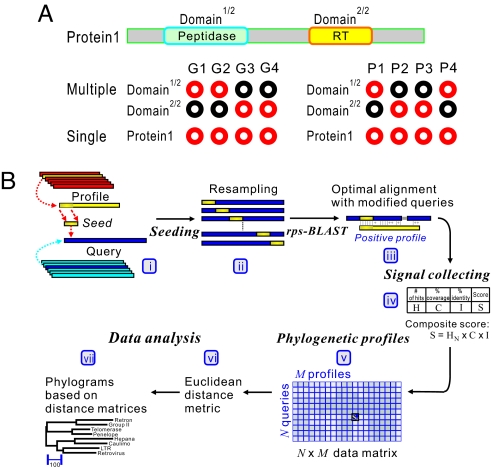
A phylogenetic profile of a protein is a vector, where each entry quantifies the existence of the protein in a different genome (35–37) (Fig. 1A Left). This approach has been shown to be applicable to whole molecule (single profile method), to an isolated domain (multiple profile method), and to individual amino acids. Further, information obtained from phylogenetic profiles allows for the generation of hypotheses about their specific functions (e.g., interaction partners), many of which are supported by existing biochemical studies. Importantly, GDDA-BLAST matrices are a variation of phylogenetic profiles. In our case, a protein is a vector where each entry quantifies the existence of alignments with a protein domain profile (Fig. 1A Right).
Fig. 1.
GDDA-BLAST concept. (A) An example illustrating binary phylogenetic profiles. Red and black circles indicate presence and absence, respectively, of a protein in a genome, or in the case of GDDA-BLAST the presence or absence of a protein alignment with a domain profile. Two methods are depicted: (i) multiple-profile method, in which individual domains in Protein 1 are individually encoded and (ii) single-profile method, in which both domains in Protein 1 are encoded together. (B) The work flow of GDDA-BLAST (see Materials and Methods). (i and ii) The algorithm begins with a modification of the query amino acid sequence at each amino acid position via the insertion of a seed sequence from the profile of interest. These seeds are obtained from the profile consensus sequences from National Center for Biotechnology Information's Conserved Domain Database (CDD). (iii–v) Signals are collected from optimal alignments between the “seeded” sequences and profiles by using rps-BLAST and are incorporated as a composite score into an N × M data matrix. (vi and vii) This data space can be analyzed to generate trees based on Euclidean distance measures and Pearson correlation measures (data not shown) of GDDA-BLAST signals, respectively.
The basic idea underlying GDDA-BLAST is to begin by compiling a set of sequence profiles with which the query sequence is compared (Fig. 1Bi) (22). These profiles can be obtained from any protein-sequence knowledge base source [e.g., Protein Data Bank, Pfam, SMART, National Center for Biotechnology Information Conserved Domain Database (CDD)] (18, 38, 39). We employ reverse specific position BLAST (rps-BLAST) (19) to detect protein domains in each query using their position specific scoring matrices (PSSMs) (39), and have introduced multiple innovations in GDDA-BLAST. We use a single domain profile database for pairwise comparisons. Then we record and quantify all alignments between an unmodified (control), and modified query sequence. To demarcate the N- and C-terminal RT domain boundaries, we used overlapping control alignments with 16 RT-specific profiles to define the regions of GDDA-BLAST measurements (see Materials and Methods and Table S2). Modified query sequences are produced by adding a standard sequence length from a profile, creating a consistent initiation site (see Materials and Methods and ref. 22). This consistent initiation site allows rps-BLAST to extend an alignment even between highly divergent sequences (22, 40–43). This strategy is designed to amplify and encode the alignments possible for any given query sequence. Moreover, it is also a way of resampling the query sequence space; instead of a sliding window, we use a sliding “seed.”
Seeds are generated from profiles by taking a portion (e.g., 3%–50%) of the profile sequence (e.g., from the N terminus, from the middle, or C terminus) (Fig. 1Bii). The variable seed length is designed to capture partial domain alignments from either direction. These seeds are inserted at each position of the query sequence. For example, a query sequence of 100 amino acids yields 100 distinct test sequences for each seed (Fig. 1Bii). A pairwise alignment for each of these modified query sequences is then measured against the parent profile by rps-BLAST (39) (Fig. 1Biii). Next, we filter our results from rps-BLAST using thresholds of number of hits, and/or percentage identity and coverage (i.e., alignment length as a function of profile length) (Fig. 1Biv). The output of these comparisons is a composite (product) score of either zero (when there is no significant match) or a positive value (which measures the degree of successful matching of the protein sequence to each of the profiles). This procedure can be readily adapted to make an unbiased comparison between a series of query sequences by subjecting them to the same screening analysis with the same set of profile sequences as those of the seeds. Thus, each query sequence is represented in a vector of nonnegative numbers in M dimensions (M = number of domain profiles tested) (Fig. 1Bv). This data can then be used to create a tree of relationships based on standard statistical techniques such as Euclidian distance between each query sequence (22) (Fig. 1Bvi–vii).
Quantitative Statistics of GDDA-BLAST Measurements.
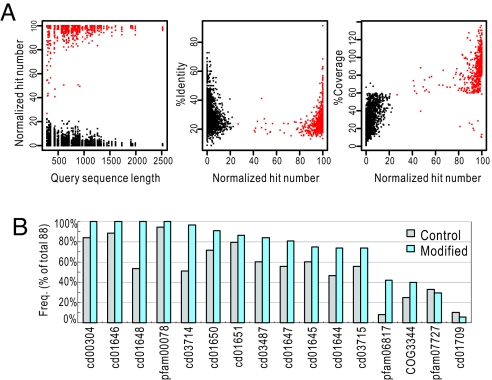
Using GDDA-BLAST, we aligned each of the unmodified 88 RT-containing sequences with all available profiles from National Center for Biotechnology Information's CDD (39). Next, we used a representative set of 43 RT sequences and 8 different seed sizes. We determined that the average number of positive profiles increases as a function of seed size, yet saturates as the seed size increases >15% (logistic fit R2 = 0.98405, data not shown). Therefore, we modified each sequence with a 10% seed size of an N- and C-terminal seed from each profile and performed high-throughput sequence alignment with the parent profile using rps-BLAST. Fig. 2 depicts quantitative statistics for all control alignments in the RT domain of 88 RT sequences. The distributions of mean percentage identity (28.46 ± 5.40 s.d.) and mean percentage coverage (77.69 ± 14.95 s.d.) are plotted as a function of normalized hits (i.e., ratio of the total number of alignments to the modified query number, scaled between 0 and 100), and the query sequence length (Fig. 2A). Importantly, we determined that alignments from the control, unmodified query do not always appear in the resampled seeded query space. These data also demonstrate that using a 60% coverage threshold removes the majority of alignments which do not improve with resampling. Therefore, resampling by seeding is one way to differentially weight alignments that can often be in similar identity/coverage ranges. Fig. 2B depicts the frequency of the 16 RT-specific profiles aligning with our dataset in control and resampled conditions. We observe that resampling increases the frequency of coverage in 14/16 cases, demonstrating that resampling can improve alignments with homologous RT profiles.
Fig. 2.
Analysis of GDDA-BLAST measurements in 88 retroelements. (A) Quantitative statistics for all control alignments in 88 RT sequences over the RT domain boundary. In these plots, we are comparing profile alignments from the control unmodified query (black) to those same profiles in the resampled seeded query (red). The distributions of query sequence length, percentage identity and percentage coverage are plotted (left to right) as a function of normalized hits (i.e., ratio of the total number of alignments to the modified query number, scaled between 0 and 100). The results indicate that resampling by seeding is one way to differentially weight alignments that can often be in similar identity/coverage ranges. (B) Bar graph depicting the frequency (% of total, n = 88) of the 16 RT-specific profiles aligning with our dataset in control (gray) and resampled (blue) conditions. The results indicate that resampling by seeding increases the frequency in 14/16 cases. This demonstrates that the consistent initiation site allows rps-BLAST to extend an alignment even between highly divergent sequences.
Performance Test of Ab Initio Evolutionary Measurements.
GDDA-BLAST.
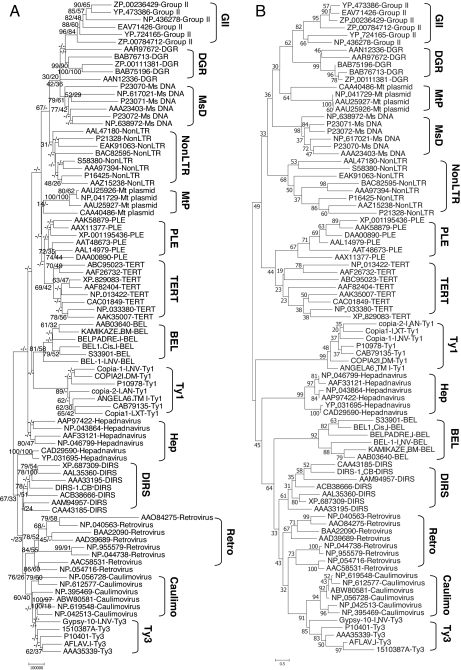
To test our hypothesis that phylogenetic profiles can derive information on genetic distance (and hence phylogenetic relationships), we quantified the Euclidean distance for each pairwise comparison in the array of 88 RT sequences from 24,280 points. Based on this measurement, an 88 × 88 distance matrix was obtained for the RT sequences and an unrooted tree was derived using the distance-based minimum evolution method available in MEGA (Fig. 3A) (44). In data not shown, we tried several different types of scoring methods (e.g., on/off binary-type score, total alignment score, composite score, weighted hit number score) and methods of phylogenetic inference [e.g., neighbor-joining, minimum evolution, Unweighted Pair Group Method with Arithmetic mean (UPGMA)]. Previously reported phylogenetic trees (23, 27, 28, 31, 45) have shown that most, if not all, of the RT groups are monophyletic. At our current settings, the tree with the highest degree of monophyly is obtained when the composite (product) score is used with the minimum evolution method. In this tree, nearly all of the 88 RT sequence display monophyly [from random considerations the probability of clustering these sequences = (1/14)88]. Importantly, we observe multiple within-group and between-group clades in our tree that are corroborated by multiple independent studies. These relationships include (i) a clear segregation of the LTR and prokaryotic clades, (ii) telomerases and PLE occurring as sister groups, and (iii) the overall topology of the LTR clade.
Fig. 3.
Phylogeny of 88 retroelements with statistical support values. (A) Unrooted phylogenetic tree of the RT domain region within 88 RT sequences derived through the estimation of evolutionary distances using GDDA-BLAST. The pairwise distances among them were acquired based on Euclidean distance measurement in the 88 × 24,280 data matrix, and an unrooted phylogenetic tree was derived from the 88 × 88 distance matrix by using a minimum evolution method. This distance measure was automatically computed without manual correction. The tree is drawn to scale, with branch lengths in the same units as those of the Euclidean distances calculated from the data matrix. Two kinds of statistical estimation for tree branching were performed and are displayed on each branch of the unrooted tree. These values are listed in the order of jackknife value and bootstrap value. The blank marked “−” in the statistical support indicates that the clustering of the branching connection cannot be measured in a standardized fashion by the given resampling method (see Materials and Methods). Bootstrap and jackknife values were obtained from 1,000 replicates and are reported as percentages. (B) Unrooted phylogenetic tree of the RT domain region within 88 RT sequences produced by multiple sequence alignment with the Dialign algorithm (default settings) and the estimation of their evolutionary distances by minimum evolution method (pairwise deletion of gaps, amino: Poisson correction, γ parameter = 1). Bootstrap values were obtained from 1,000 replicates and are reported as percentages. For both A and B, the individual groups are labeled with a bracket. The major differences between these two trees lie in the statistical support values and the placement of the Mt plasmids, hepadnaviruses, and LTR BEL subgroup.
We tested the robustness of the GDDA-BLAST phylogeny by two types of resampling methods: bootstrap and jackknife, each with 1,000 replicates. These statistical support values are displayed in order of jackknife and bootstrap in our tree (Fig. 3A). Despite no measurable statistical support at some nodes, several noticeable features are found in the statistical values to support reliable branching patterns.
Multiple sequence alignment.
As comparison we performed multiple sequence alignment of 88 RT domains (as defined by GDDA-BLAST) with various established algorithms that measure evolutionary distance [e.g., ClustalW (46), MUSCLE (47, 48), K-align (4, 49), and Dialign (50)]. From each of the resulting alignments we inferred a minimum evolution tree (see Materials and Methods and Fig. S2) that includes no manual editing. When trees are constructed by using complete deletion of gaps, none of these methods provide a monophyletic tree (data not shown). When we perform our analyses with pairwise deletion of gaps, both ClustalW and Dialign obtain a monophyletic tree. The tree generated with Dialign obtains the best statistical support of all methods tested and is very similar to the one generated by GDDA-BLAST in both monophyly and topology.
Discussion
Our case study using a benchmark phylogeny in the twilight zone of sequence similarity demonstrates that phylogenetic profiles are capable of inferring deep evolutionary relationships. Thus, we conclude that phylogenetic profiles generated using profile-sequence alignments below statistical thresholds are not necessarily random and can retain biologically relevant information that is not reflected by chance similarity.
It is important to compare the independent results obtained by GDDA-BLAST and Dialign with results in the literature obtained by manual editing of retroelement sequences. Based on random considerations, obtaining similar results for the 14 clades of retroelements tested with GDDA-BLAST and Dialign is grossly improbable. For example, non-LTR and LTR elements are clearly distinct, as has been suggested previously (27, 33). Telomerases and PLEs also form a sister clade as reported by Arkhipova and Doulatov (28, 33). In large part, both phylogenies also recapitulate the results of Goodwin and Poulter for the topology of the LTR group, including retroviruses and pararetroviruses (32). With the exception of Mt plasmids, the topology for the prokaryotic group is the same in both analyses, which accord with previous studies (23, 28, 33). All MSA methods tested here and manually edited trees in the literature place the Mt plasmids in the prokaryotic group. Conversely, GDDA-BLAST places Mt plasmids with the telomerases and PLEs, although this position has no statistical support. Nevertheless, it has been demonstrated that Mt plasmids have 3′ terminal repeats similar to those of chromosomal telomeres, making them the potential precursor of telomerase (51). Another key difference between the Dialign and GDDA-BLAST results is the placement of the hepadnaviruses. Dialign places the hepadnaviruses between the Bel/Ty1 clades, in contrast to the results obtained with GDDA-BLAST and previous reports (32, 52).
Within the twilight zone of sequence similarity, statistical support can help eliminate evolutionary ambiguities. Although none of the ab initio methods tested here obtained robust deep-branch statistical support, having an independent approach to estimate evolutionary relationships undoubtedly represent an important advance (Fig. 3). Determining the data points collected by GDDA-BLAST that are informative for SF&E annotation should enable us to optimize and refine our approach. Evidence for this idea is shown in Fig. S3. In these analyses, we limited our measurements to the profiles in the control preparation that are “active” (defined by having a normalized hit ratio >25). When we generated trees from both the unmodified and modified sequences for these profiles, we observed that even this limited subset of profiles have significant informational content. When we further limited this tree to only the most frequently occurring alignments, we obtained the 16 RT domain-containing profiles present in the CDD. Intriguingly, we still observe significant monophyly that is well above random. Based on these results, it is reasonable to consider that expanding only the informative profiles within our knowledge base will improve the robustness of GDDA-BLAST measurements, in addition to improving computational performance.
Despite efforts by multiple investigators over 20 years, there still is no commonly accepted phylogenetic history of the retroelements. This is due, in large part, to the extreme divergence between the sequences in question, which compromises both multiple sequence alignment and phylogenetic analyses. GDDA-BLAST uses knowledge-based screens of SF&E domain profiles. Therefore, these independent measurements derive additional information toward characterizing the evolutionary history of proteins that cannot be easily obtained from multiple sequence alignment methods. Even in its nascent stage, GDDA-BLAST provides measurements that give independent support for phylogenetic studies and key insight into evolutionary relationships among distantly related and/or rapidly evolving proteins.
Materials and Methods
Sequence Collection.
A total of 88 RT sequences were collected from GenBank and the Genetic Information Research Institute Repbase (http://www.girinst.org), representing a sample of the known diversity of RT-containing organisms (see Table S1; those curated from Repbase are denoted as TRACE).
Defining RT Domain Boundaries.
We performed quantitative statistics of the overlapping alignments for 16 RT-specific profiles (extracted from CDD) in each of the 88 unmodified RT sequences. The start and end position were defined from the start of the most N-terminal alignment and the end of the most C-terminal alignment. Statistics for these alignments are reported in Table S2 and include (i) position of the alignments; (ii) the percentage identity and coverage of each alignment; (iii) the frequency (% of total) of each domain profile aligning in the total dataset of 88 RT sequences; and (iv) a list of the 16 RT profiles along with their identifier, length, and description.
GDDA-BLAST Phylogenetic Trees.
Each RT was screened in GDDA-BLAST with 24,280 profiles at 10% seed size. For each profile scoring above threshold (60% coverage, 10% identity), a composite score was generated (%coverage × %identity × normalized hit number), creating an N (queries) × M (profile) matrix. Next, the Euclidian distance for the product scores of each query sequence was calculated, creating an N (queries) × N (Euclidian distance) matrix. These distances matrices can then be used to infer phylogenetic trees using an appropriate method (minimum evolution, neighbor joining, UPGMA, etc). See SI Materials and Methods for a complete description.
Statistical Estimation of Phylogenetic Trees.
Two different statistical support values are provided on the branches of our best phylogenetic tree, which we obtained with 88 sequences and 24,280 domain profiles at 10% seed size. All branches in the tree are not necessarily supported by all of the statistical tests we performed. For each sample, we generated a minimum evolution tree and obtained a consensus tree of the trees. Given the consensus tree, we provided statistical support values on branches of our best phylogenetic tree if the branches were shared between the original tree and the tree obtained from the statistical test. See SI Materials and Methods for a complete description of all methods used.
Supplementary Material
Acknowledgments.
We thank Jason Holmes at the Pennsylvania State University CAC center for technical assistance; Drs. Jayanth Banavar and Peter Hudson for useful discussion and continued support; and Jim White, Richard Wilhelm, M. Yahu, J.L. Children, C. Diddy, and Barbara VanRossum for creative dialogue. This work was supported by the Searle Young Investigators Award and start-up monies from Pennsylvania State University (R.L.P.), NCSA Grant TG-MCB070027N to R.L.P. and D.V.R., Pittsburgh Supercomputing Center Grant BIO060003P to R.L.P., and a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds to D.V.R. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803860105/DCSupplemental.
References
- 1.Ponting CP, Russell RR. The natural history of protein domains. Annu Rev Biophys Biomol Struct. 2002;31:45–71. doi: 10.1146/annurev.biophys.31.082901.134314. [DOI] [PubMed] [Google Scholar]
- 2.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Park J, et al. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J Mol Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- 4.Lassmann T, Sonnhammer EL. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 6.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 7.Socolich M, et al. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 8.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blake JD, Cohen FE. Pairwise sequence alignment below the twilight zone. J Mol Biol. 2001;307:721–735. doi: 10.1006/jmbi.2001.4495. [DOI] [PubMed] [Google Scholar]
- 12.Taylor WR. Identification of protein sequence homology by consensus template alignment. J Mol Biol. 1986;188:233–258. doi: 10.1016/0022-2836(86)90308-6. [DOI] [PubMed] [Google Scholar]
- 13.Yi TM, Lander ES. Recognition of related proteins by iterative template refinement (ITR) Protein Sci. 1994;3:1315–1328. doi: 10.1002/pro.5560030818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gribskov M, McLachlan AD, Eisenberg D. Profile analysis: Detection of distantly related proteins. Proc Natl Acad Sci USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luthy R, Xenarios I, Bucher P. Improving the sensitivity of the sequence profile method. Protein Sci. 1994;3:139–146. doi: 10.1002/pro.5560030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elofsson A, Fischer D, Rice DW, Le Grand SM, Eisenberg D. A study of combined structure/sequence profiles. Fold Des. 1996;1:451–461. doi: 10.1016/S1359-0278(96)00061-2. [DOI] [PubMed] [Google Scholar]
- 17.Baldi P, Chauvin Y, Hunkapiller T, McClure MA. Hidden Markov models of biological primary sequence information. Proc Natl Acad Sci USA. 1994;91:1059–1063. doi: 10.1073/pnas.91.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnhammer EL, Eddy SR, Durbin R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneidman E, Berry MJ, Segev R, Bialek W. Weak pairwise correlations imply strongly correlated network states in a neural population. Nature. 2006;440:1007–1012. doi: 10.1038/nature04701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko KD, et al. Phylogenetic profiles as a unified framework for measuring protein structure, function and evolution. Phys Arch. 2008 arXiv:0806.239, q-bio.Q. [Google Scholar]
- 23.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeke JD. The unusual phylogenetic distribution of retrotransposons: A hypothesis. Genome Res. 2003;13:1975–1983. doi: 10.1101/gr.1392003. [DOI] [PubMed] [Google Scholar]
- 25.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 26.Darnell JE, Doolittle WF. Speculations on the early course of evolution. Proc Natl Acad Sci USA. 1986;83:1271–1275. doi: 10.1073/pnas.83.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arkhipova IR, Pyatkov KI, Meselson M, Evgen'ev MB. Retroelements containing introns in diverse invertebrate taxa. Nat Genet. 2003;33:123–124. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- 29.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 31.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 32.Goodwin TJ, Poulter RT. Multiple LTR-retrotransposon families in the asexual yeast Candida albicans. Genome Res. 2000;10:174–191. doi: 10.1101/gr.10.2.174. [DOI] [PubMed] [Google Scholar]
- 33.Doulatov S, et al. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 34.Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y, Subramaniam S. Locally defined protein phylogenetic profiles reveal previously missed protein interactions and functional relationships. Proteins. 2006;62:1115–1124. doi: 10.1002/prot.20830. [DOI] [PubMed] [Google Scholar]
- 36.Ranea JA, Yeats C, Grant A, Orengo CA. Predicting protein function with hierarchical phylogenetic profiles: The Gene3D Phylo-Tuner method applied to eukaryotic genomes. PLoS Comput Biol. 2007;3:e237. doi: 10.1371/journal.pcbi.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letunic I, et al. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchler-Bauer A, et al. CDD: A conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rossum DB, et al. Phospholipase Cgamma1 controls surface expression of TRPC3 through an intermolecular PH domain. Nature. 2005;434:99–104. doi: 10.1038/nature03340. [DOI] [PubMed] [Google Scholar]
- 41.Caraveo G, van Rossum DB, Patterson RL, Snyder SH, Desiderio S. Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science. 2006;314:122–125. doi: 10.1126/science.1127815. [DOI] [PubMed] [Google Scholar]
- 42.Nikolaidis N, et al. Ancient origin of the new developmental superfamily DANGER. PLoS ONE. 2007;2:e204. doi: 10.1371/journal.pone.0000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraborty A, et al. HSP90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc Natl Acad Sci USA. 2008;105:1134–1139. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lassmann T, Sonnhammer EL. Kalign, Kalignvu and Mumsa: Web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–W599. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgenstern B. DIALIGN: Multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 2004;32:W33–6. doi: 10.1093/nar/gkh373. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walther TC, Kennell JC. Linear mitochondrial plasmids of F. oxysporum are novel, telomere-like retroelements. Mol Cell. 1999;4:229–238. doi: 10.1016/s1097-2765(00)80370-6. [DOI] [PubMed] [Google Scholar]
- 52.Plant EP, Goodwin TJ, Poulter RT. Tca5, a Ty5-like retrotransposon from Candida albicans. Yeast. 2000;16:1509–1518. doi: 10.1002/1097-0061(200012)16:16<1509::AID-YEA638>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.