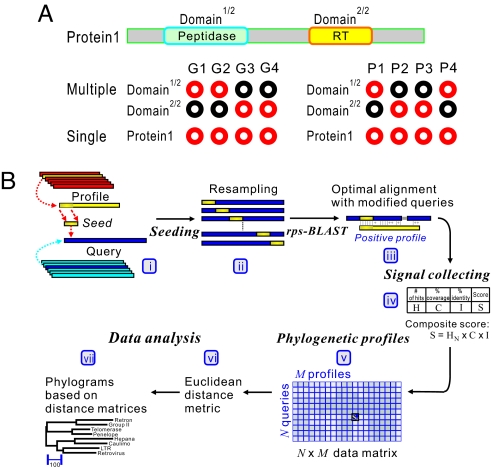
Fig. 1.
GDDA-BLAST concept. (A) An example illustrating binary phylogenetic profiles. Red and black circles indicate presence and absence, respectively, of a protein in a genome, or in the case of GDDA-BLAST the presence or absence of a protein alignment with a domain profile. Two methods are depicted: (i) multiple-profile method, in which individual domains in Protein 1 are individually encoded and (ii) single-profile method, in which both domains in Protein 1 are encoded together. (B) The work flow of GDDA-BLAST (see Materials and Methods). (i and ii) The algorithm begins with a modification of the query amino acid sequence at each amino acid position via the insertion of a seed sequence from the profile of interest. These seeds are obtained from the profile consensus sequences from National Center for Biotechnology Information's Conserved Domain Database (CDD). (iii–v) Signals are collected from optimal alignments between the “seeded” sequences and profiles by using rps-BLAST and are incorporated as a composite score into an N × M data matrix. (vi and vii) This data space can be analyzed to generate trees based on Euclidean distance measures and Pearson correlation measures (data not shown) of GDDA-BLAST signals, respectively.