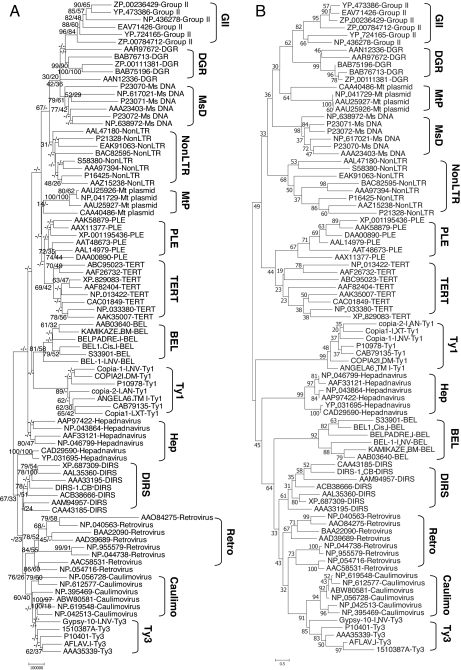
Fig. 3.
Phylogeny of 88 retroelements with statistical support values. (A) Unrooted phylogenetic tree of the RT domain region within 88 RT sequences derived through the estimation of evolutionary distances using GDDA-BLAST. The pairwise distances among them were acquired based on Euclidean distance measurement in the 88 × 24,280 data matrix, and an unrooted phylogenetic tree was derived from the 88 × 88 distance matrix by using a minimum evolution method. This distance measure was automatically computed without manual correction. The tree is drawn to scale, with branch lengths in the same units as those of the Euclidean distances calculated from the data matrix. Two kinds of statistical estimation for tree branching were performed and are displayed on each branch of the unrooted tree. These values are listed in the order of jackknife value and bootstrap value. The blank marked “−” in the statistical support indicates that the clustering of the branching connection cannot be measured in a standardized fashion by the given resampling method (see Materials and Methods). Bootstrap and jackknife values were obtained from 1,000 replicates and are reported as percentages. (B) Unrooted phylogenetic tree of the RT domain region within 88 RT sequences produced by multiple sequence alignment with the Dialign algorithm (default settings) and the estimation of their evolutionary distances by minimum evolution method (pairwise deletion of gaps, amino: Poisson correction, γ parameter = 1). Bootstrap values were obtained from 1,000 replicates and are reported as percentages. For both A and B, the individual groups are labeled with a bracket. The major differences between these two trees lie in the statistical support values and the placement of the Mt plasmids, hepadnaviruses, and LTR BEL subgroup.