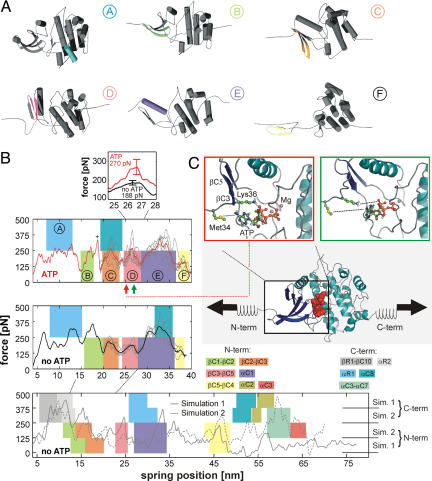
Fig. 3.
Molecular dynamics (MD) simulations of the force-induced unfolding of titin kinase (TK). (A) Representative unfolding intermediates with the unfolding secondary structure elements colored according to the scheme in Fig. 1D; the β-strands unfold pairwise, and colors refer to the respective N-terminal strand of each pair. (B) Unfolding forces of truncated TK with ATP (Top), without ATP (Middle), and of the complete TK (Bottom). For the complete TK, two independent 90-ns simulations were carried out (solid and dashed lines). Starting from a partially unfolded structure at ≈19 nm, five 26-ns trajectories (thin gray lines) were averaged for both sets of simulations (thick lines in Top and Middle). Color-shaded areas indicate main unfolding events, which correspond to the colors used in A and in Fig. 1D. An additional force peak in the presence of ATP is predicted (plus sign and pink-shaded area in Top). This force peak (Inset) is higher for bound ATP (270 pN) than for an empty binding pocket (188 pN). Because of the necessarily much faster pulling rates of 0.8 m/s used for the simulations, larger unfolding forces are seen, which can be related to the experimental loading rates (11). (C) In the force–probe MD simulations, harmonic springs were attached to the protein and retracted with constant velocity (lower schematic, ATP shown as red spheres). (C Insets) Representative structures shortly before (Left) and after (Right) the ATP force peak. ATP and the two key residues methionine-34 and lysine-36 are shown in ball-and-stick representation, and the rupture of molecular interactions is indicated by dotted lines.