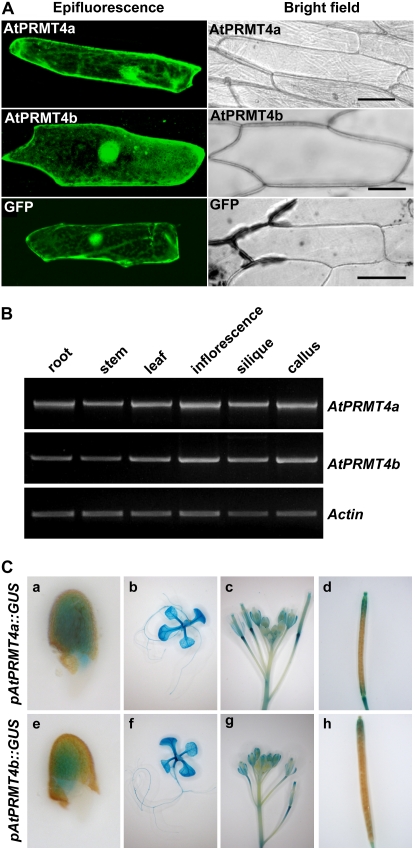
Figure 3.
AtPRMT4a and AtPRMT4b exhibited similar subcellular distribution and expression patterns. A, AtPRMT4a and AtPRMT4b cDNAs were fused in-frame to the C-terminal end of GFP. The resulting constructs were transiently transformed into onion epidermal cells by DNA-coated gold particle bombardment, and protein localization was observed by confocal microscopy, shown as epifluorescence and bright-field images. Bars = 75 μm. B, Semiquantitative RT-PCR was performed to examine the spatial expression of AtPRMT4a and AtPRMT4b in root, stem, leaf, inflorescence, silique, and callus. Actin2/7 was used as a constitutive positive control. C, Histochemical GUS activity analysis of pAtPRMT4a∷GUS and pAtPRMT4b∷GUS transgenic plants in the Col-0 background. a and e, Germinating seeds; b and f, 10-d-old-seedlings; c and g, inflorescences; d and h, siliques.