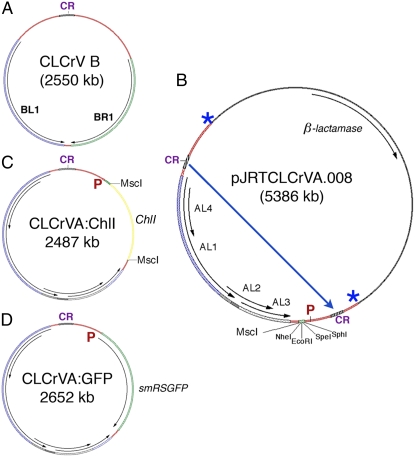
Figure 1.
Plasmids used for bombardment. A, Wild-type CLCrV B component showing divergent transcription of the BR1 and BL1 genes from the common region (CR). The CLCrV DNA-A component (not shown) has a similar coding region and five divergently transcribed genes. Both components are needed for systemic infection. B, E. coli plasmid containing the A component of the empty VIGS vector. The coat protein (AR1) open reading frame has been replaced with an MCS with MscI, NheI, EcoR1, SpeI, and SphI recognition sites for cloning gene silencing fragments behind the AR1 promoter and before the putative polyA site. The duplicated common regions in this plasmid allow replicational release of unit length episomes, such as in A, C, and D, following introduction into plant cells. The long blue arrow shows sites in the coding region that are nicked by the AL1 Rep protein for initiation of rolling circle replication and then AL1-mediated ligation to circularize the viral DNA. Overlapping arrows in the viral DNA show AL1, AL4, AL2, and AL3 genes. E. coli sequence has the β-lactamase gene conferring ampicillin resistance. P, AR1 promoter. Asterisks show the junctions between viral and plasmid DNA. C, CLCrV A component of the VIGS vector for silencing ChlI. It is the same as the viral DNA part of the plasmid shown in B except that a 500-bp gene fragment from the cotton ChlI gene has been cloned into the MscI site for transcription by the AR1 promoter (P). This episome contains a single common region and is small enough to move between cells. D, CLCrV A component used as an expression vector for rsSMGFP. It is the same as C except that a full-length smRSGFP gene has been cloned into the MCS in sense orientation with respect to the AR1 promoter (P). [See online article for color version of this figure.]