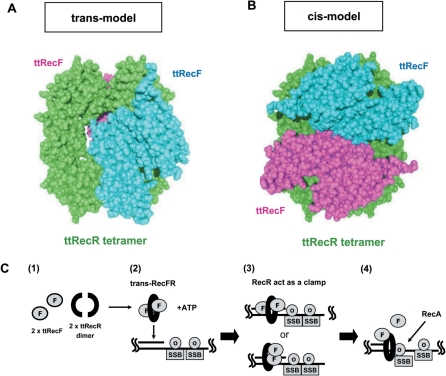
Figure 5.
A rigid body model structure of the ttRecFR complex and proposed scheme of its function. (A) A trans model of the ttRecFR complex constructed by the rigid body model method. Two ttRecF monomers (cyan and pink) bind to the opposite side of the ttRecR tetramer (green). (B) A cis model of the ttRecFR complex constructed by the rigid body model method. Two ttRecF monomers bind to the same side of the ttRecR tetramer. (C) Schematic diagram of the dsDNA clamp model of the ttRecR tetramer in ttRecFOR assembly at dsDNA–ssDNA junctions. dsDNA–ssDNA junctions are generated as a consequence of DNA damage. (1) Two ttRecR dimers form a ring-like tetramer with two ttRecF monomers, resulting in a trans-RecFR complex. (2) ATP causes large conformational changes of the ttRecFR complex to tether the complex to dsDNA. (3) A trans (above) or cis (below) ttRecFR complex recognizes or binds to the junction site and then interacts with ttRecO localized on ssDNA regions containing the SSB protein. (4) The ttRecOR complex, which may associate with ttRecF, facilitates the loading of RecA onto SSB-coated ssDNA. DNA damage is subsequently repaired by homologous recombination mediated by RecA.