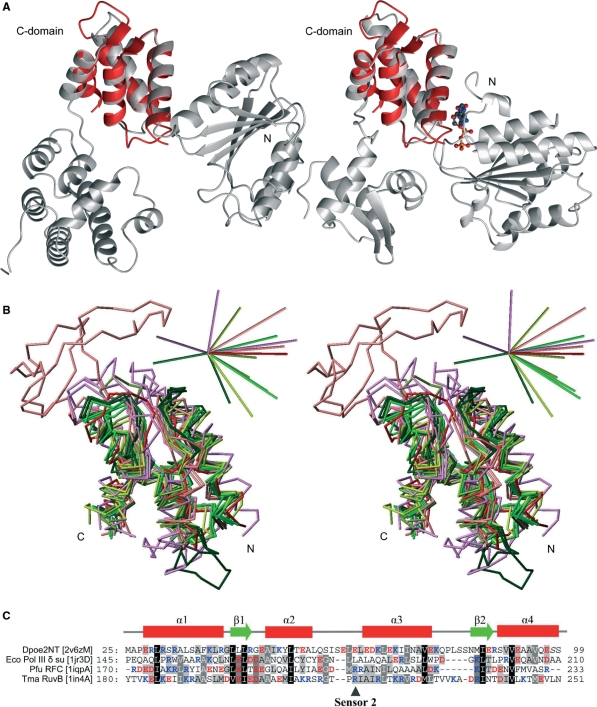
Figure 4.
Comparison of the amino terminal structure of Pol ε subunit B with C-domain structures of AAA+ family proteins. (A) Ribbon presentation of the δ subunit of E. coli Pol III γ complex (PDB entry 1jr3D) on the left and the T. maritime RuvB Holliday junction branch migration motor protein with bound ADP molecule (PDB entry 1in4A) on the right. The Dpoe2NT solution structure superimposed on the C-domain is represented in red. The ADP molecule present in structure 1in4A was included to emphasize the location of the homologous structure relative to the functional site at the domain interphase. (B) Superimposed Cα traces of C-domain structures against Dpoe2NT. The color scheme was adopted from the coloring in the cluster maps by Ammelburg and co-workers (43): TmaRuvB (PDB ID 1in4A), Tth RuvB (1hqcA) and Hsa RuvBL1 (2c9oA) are in bright green, crenarchaeal replication initiator proteins Ape ORC2 (1w5sA) and Pae Cdc6 (1fnnA) are in dark green, Eco Pol III clamp loader subunits δ (1jqjC) and gamma (1jr3A) are in greenish yellow, while clamp loader proteins from Pfu (RFCS, 1iqpA) and yeast (RFC3, 1sxjC) are in olive green. Bacterial proteases and chaperones Bsu Lon1 (1x37A), Eco ClpA (1r6bX) and Eco HslU (1do0A) are in violet, and proteins belonging to AAA-D1 cluster, Hsa VPS4B (1xwiA) and Tth Ftsh (2dhrA) are in light red. Dpoe2NT (2v6zM) is in red. 3D bars at upright in stereo view indicate directions (positive) and relative magnitude (0.57 debye/atom on average) of calculated dipole moments for each domain structures independently. (C) Structure-based sequence alignment of a representative selection of homological C-domain structures: E. Coli Pol III γ complex subunit δ (1jr3D), Pyrococcus furiosus Clamp loader small subunit (1iqpA) and T. maritime RuvB Holliday junction branch migration motor protein (1in4A) against Dpoe2NT (2v6zM). Secondary structure elements of Dpoe2NT are indicated above the alignment.