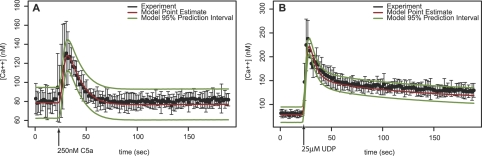
Figure 3. Model simulations are compared to experimental data.
The point estimate is computed using the posterior distribution of the parameter as estimated by Markov chain Monte Carlo given the data from 96 experiments on C5a and UDP at various doses in combination with 5 different shRNAi knockdown cell lines. The 95% posterior predictive intervals are estimated by Monte Carlo simulations including both parameter and measurement uncertainty. The measured mean and approximate 95% confidence intervals of four replicates is shown by a black dot and error bar. (A) C5a at 250 nM was introduced at 20s and the experimentally observed pulse in cytosolic calcium concentration is shown. (B) The qualitative shape of the calcium pulse for 25 µM UDP is different than for 250 nM C5a. The pulse does not completely adapt and return to the prestimulated level. For both ligands, the model prediction confidence intervals overlap the data error bars that indicate the model fit is consistent with the data within the measurement uncertainty.