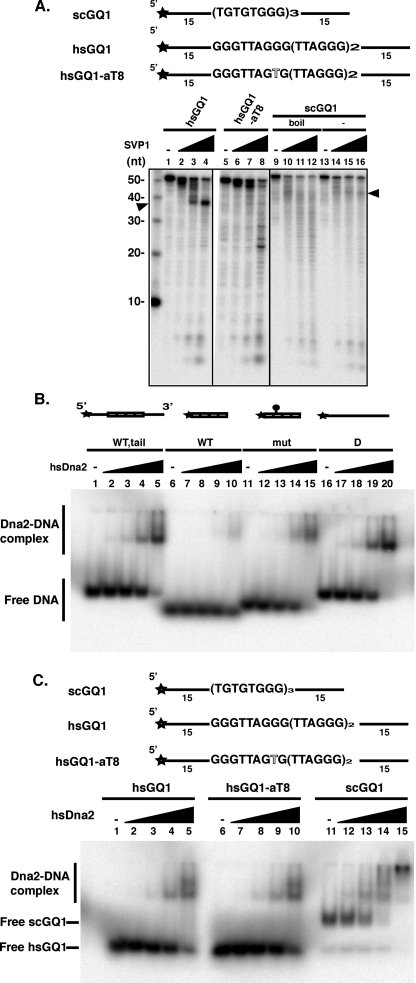
FIGURE 6.
Characterization of G4 DNA formed by hsGQ1, mutant hsGQ1-aT8, and scGQ1 determined by sensitivity to SVPI. A, formation of G4 DNA is inhibitory to SVPI nuclease. 50 fmol of 5′ end-labeled substrates of human wild type (hsGQ1) and mutant (hsGQ1-aT8) telomeric substrates were treated with SVPI nuclease as described under”Experimental Procedures.“Nuclease products were analyzed by denaturing gel electrophoresis. Substrates were preincubated in the absence (lanes 1 and 5) or the presence of increasing amounts of SVPI nuclease as follows: 75 ng (lanes 2 and 6), 150 ng (lanes 3 and 7), and 300 ng (lanes 4 and 8) of G4 DNA was formed from the hsGQ1 oligonucleotides by boiling and slow cooling. hsGQ1-aT8 contains a G4-disrupting mutation in the second G-tract. Wedges represent increasing amounts of SVPI nuclease; – represents no enzyme addition. Arrowhead points to the band generated by removal of the 15-nucleotide 3′ tail by SVPI and the block at the junction with G4 DNA. Left lane shows length markers (nt). Lanes 9–16, scGQ1 G4 DNA was formed as described under”Experimental Procedures.“Single-stranded scGQ1 was prepared by boiling the G4 DNA. Lanes 13–16 show pausing of SVPI as the nuclease encounters the G4 DNA from the 3′ direction. Lanes 9–12 show cutting in the G-rich sequences (bands less than 40 nt in length). B, hsDna2 needs single-stranded region for efficient binding to hsGQ1 G4 DNA. Reaction mixtures contained 15 fmol of hsGQ1 wild type with tail (lanes 1–5), hsGQ wild type without tail (lanes 6–10), hsGQ mutant without tail (lanes 11–15), and single-stranded DNA (lanes 16–20) substrates and were incubated with hsDna2 at 0, 5, 25, 100, and 690 fmol (represented as triangles above lanes 1–5, 6–10, 11–15, and 16–20). Formation of protein-DNA complex was analyzed by gel shift assay. C, comparison of hsDna2 binding to yeast (intermolecular) and human (intramolecular) G4. Reaction mixtures contained 15 fmol of hsGQ1 wild type (lanes 1–5), hsGQ1-aT8 mutant (lanes 6–10, G4), and scGQ1 (lanes 11–15) substrates and were incubated with hsDna2 at 0, 5, 25, 100, and 690 fmol (represented as triangles above lanes 1–5, 6–10, and 11–15). Formation of protein-DNA complex was analyzed by gel shift assay.