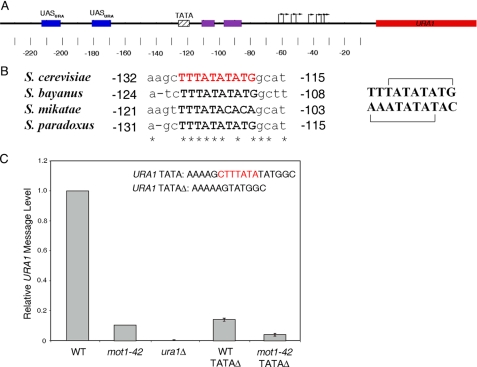
FIGURE 1.
Organization of the URA1 promoter and requirement for the URA1 TATA element in vivo. A, schematic of the URA1 locus and 5′-untranslated region showing the locations of regulatory regions identified through prior molecular studies and phylogenetic sequence alignments. The location of the TATA box is indicated by the hatched box, and the transcription start sites (43) are shown by the arrows. The URA1 promoter harbors two binding sites for the Ppr1 transcription factor (52, 53) (blue boxes, UASURA), as well as two conserved sequences (purple boxes) that may be also be involved in URA1 transcriptional control (43). The URA1 gene product is required for uracil biosynthesis (43). In our strain background, Ppr1 is required for URA1 expression; however, transcriptional activity is insensitive to the level of uracil in the media (data not shown). Numbers indicate position relative to the start of translation, ATG. B, sequence alignment showing the TATA element of URA1 in four yeast strains. Note the S. cerevisiae“conventional” TATA element TATATATG and the overlapping reverse TATA element TTTATATA (shown in red). Asterisks indicate identical base pairs, and numbers are relative to the start of translation. The top and bottom strands of the S. cerevisiae sequence are shown on the right, with the overlapping putative TATA elements marked by brackets. C, quantitation of URA1 message levels in the indicated strains, determined by Northern blotting. The 7-bp region shown in red (top right) was deleted in the TATAΔ strains. URA1 RNA levels were normalized to the levels of ACT1 RNA in the same samples. Errors are standard deviations obtained from three separate RNA samples for each strain.