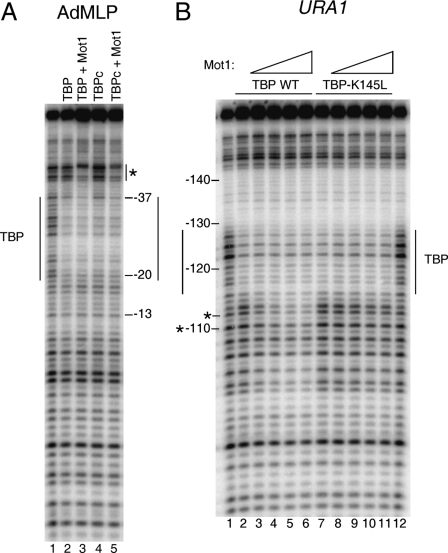
FIGURE 3.
Organization of the Mot1-TBP-URA1 DNA complex. A, DNase I footprinting analysis of TBP and Mot1 binding to the radiolabeled AdMLP probe containing the TATA element TATAAAAG. The reactions contained no added protein (1st lane), full-length TBP (14 nm, 2nd and 3rd lanes), TBP core domain (TBPc, 8 nm, 4th and 5th lanes), and 11.6 nm Mot1 (3rd and 5th lanes). Reactions were incubated at 22 °C for ∼20 min and then processed as described previously (31). The TBP footprint is indicated by the long vertical lines, and upstream protection induced by Mot1 binding is shown by the asterisk. B, DNase I footprinting experiment in which Mot1 was added to TBP-URA1 DNA complexes. DNA and 14 nm TBP (WT or K145L) were incubated together for ∼20 min at 22 °C, followed by the addition of Mot1 as follows: 1.2 nm to lanes 3 and 8; 3.6 nm to lanes 4 and 9; 7.2 nm to lanes 5 and 10; and 10.8 nm to lanes 6 and 11. After ∼5 min of incubation, reactions were processed as described (31). Lanes 1 and 12 show free DNA. Vertical lines indicate the TBP footprint, and asterisks indicate downstream positions where digestion was inhibited by Mot1 below the level of digestion seen in DNA alone (see Fig. 4).