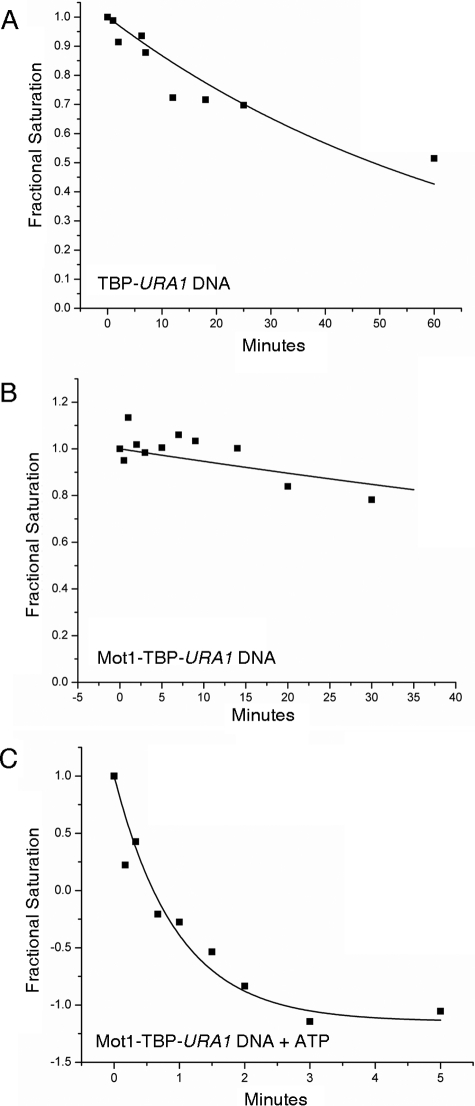
FIGURE 8.
Dissociation kinetic analysis of complexes formed on the URA1 promoter. Radiolabeled URA1 DNA probe was incubated with 14 nm TBP with or without 15 nm Mot1. At time 0, an excess of unlabeled TATA DNA was added to the reactions, and the extent of TATA box occupancy was determined by DNase I footprinting as described previously (31). A, rate of dissociation of the TBP-URA1 DNA complex. The half-time value for the best fit to a single exponential decay is 70.4 ± 8.3 min. The TBP-DNA complexes formed on the URA1 template are about 4-fold more stable than those formed on the AdMLP (31) (supplemental Table S1). B, rate of dissociation of the Mot1-TBP-URA1 DNA complex. The half-time value for the best fit to a single exponential is 181.9 ± 67.2 min. C, rate of dissociation of the Mot1-TBP-URA1 DNA complex in the presence of 25 μm ATP, which was added at time 0. The half-time value for the best fit to a single exponential is 0.96 ± 0.11 min. This represents an approximate 190-fold increase in the dissociation rate as a result of Mot1 ATP-dependent catalytic activity.