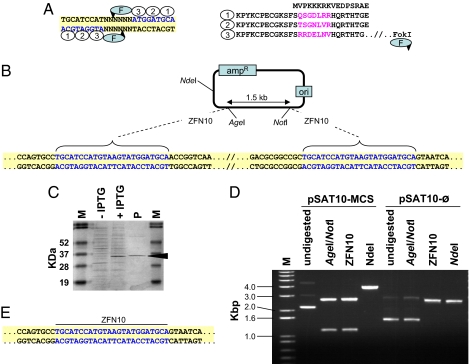
Fig. 1.
Structure and expression of ZFNs and their use for DNA cloning. (A) The structure of a typical 24-bp-long ZFN recognition site and its corresponding zinc finger protein, exemplified by ZFN10. DNA triplets (blue) and their binding zinc fingers (ovals) are numbered correspondingly. The unique amino acid sequences in each zinc finger are in purple. The FokI cleavage domain (F) is linked to the zinc finger protein's carboxyl terminus, and the predicted cleavage sites are indicated by arrowheads. (B) Scheme of the pSAT10-MCS plasmid. Sequences of the ZFN10 palindrome-like recognition sites are in blue. (C) Separation of total crude extract from induced (+IPTG) and noninduced (-IPTG) ZFN10 protein-expressing E. coli cells and of purified ZFN10 protein (P). The 34-kDa band corresponding to the ZFN10 protein is indicated. (D) Restriction analysis of the parental pSAT10-MCS and its progeny plasmid pSAT10- . (E) Sequence analysis of the ZFN10 ligation site in one of the pSAT10-
. (E) Sequence analysis of the ZFN10 ligation site in one of the pSAT10- plasmids. Sequences of the reconstructed ZFN10 palindrome-like recognition sites are in blue.
plasmids. Sequences of the reconstructed ZFN10 palindrome-like recognition sites are in blue.