Abstract
Primary cilia are present on mammalian neurons and glia, but their function is largely unknown. We generated conditional homozygous mutant mice for a gene we termed Stumpy. Mutants lack cilia and have conspicuous abnormalities in postnatally developing brain regions, including a hypoplasic hippocampus characterized by a primary deficiency in neural stem cells known as astrocyte-like neural precursors (ALNPs). Previous studies suggested that primary cilia mediate sonic hedgehog (Shh) signaling. Here, we find that loss of ALNP cilia leads to abrogated Shh activity, increased cell cycle exit, and morphological abnormalities in ALNPs. Processing of Gli3, a mediator of Shh signaling, is also altered in the absence of cilia. Further, key mediators of the Shh pathway localize to ALNP cilia. Thus, selective targeting of Shh machinery to primary cilia confers to ALNPs the ability to differentially respond to Shh mitogenic signals compared to neighboring cells. Our data suggest these organelles are cellular “antennae” critically required to modulate ALNP behavior.
Keywords: adult neurogenesis, Gli1, Patched, Smoothened, stem cell
Cilia are conserved microtubule-based organelles that grow from basal bodies (a centrosome-derived structure) and protrude from the cell surface (1). A growing number of biological processes and clinical disorders are attributable to properly functioning or dysfunctional cilia, respectively (1–4). Neurons and astrocytes have been found to harbor a nonmotile, primary cilium (5, 6). Despite this widespread presence of primary cilia in the forebrain, their function remains elusive (6).
Recent findings suggest that the Shh signaling pathway in vertebrates appears to be mediated through the primary cilium (4, 7, 8). Whether this pathway is active in primary cilia in the postnatal brain is unknown. Shh signaling is a key regulator of cell proliferation in the external granule cell layer (EGL) of the cerebellum, dentate gyrus (DG) of the hippocampus, and subependymal zone (SEZ) of the lateral ventricles (9, 10): three brain regions that continue to develop postnatally (11). Notably, it was demonstrated that Shh-responsive postnatal precursors appear to be a unique population that are derived relatively late in embryogenesis (10, 12, 13). The mechanisms governing this postnatal responsiveness to Shh are unclear. Therefore, we hypothesize that this responsiveness may occur through cilia within germinal regions of the postnatal brain.
Results
Stumpy Is Required for Development of Postnatal Brain Structures and Ciliogenesis.
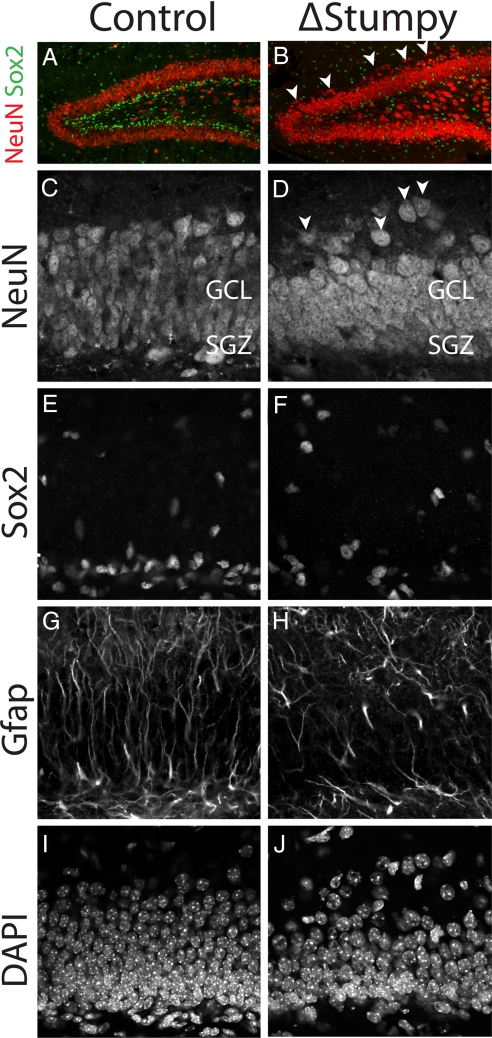
During characterization, a conditional mutant mouse for the Stumpy gene, we observed gross olfactory bulb (data not shown), hippocampal, and cerebellar abnormalities in P13 mice [Fig. 1, supporting information (SI) Fig. S1]. In particular, the DG granule cell layer was thinner when compared to control mice and showed dispersion of NeuN+ granule cells (Figs. 1 A–D and Fig. S2). When the underlying glial scaffold was observed by glial fibrillary acidic protein (Gfap) immunoreactivity, we noted that the typical lamination and morphology of glial cells was severely disrupted (Fig. 1 E–H). Glial cell bodies, as shown by combined Sox2 and Gfap immunoreactivity, were located more randomly in NestinCre; Stumpyfl/fl mice (heretofore referred to as ΔStumpy mice), and there was a notable lack of DG radial glia (Fig. 1 A, B, and E–H).
Fig. 1.
Gross hippocampal defects in Stumpy-deficient brain. (A and B) Immunostaining at P13 for NeuN revealed an overall smaller granule cell layer (GCL) and dispersed neurons (white arrowheads) in Stumpy-mutant (ΔStumpy) compared to control brains. (C and D) NeuN-labeled neuronal nuclei showed reduced thickness of the GCL in mutant (D) compared to heterozygous littermates (C). White arrowheads denote dispersed granule cells. (E and F) Sox2-labeled nuclei were dispersed in ΔStumpy (F) compared to control (E). (G and H) GFAP immunostaining in ΔStumpy brain (H) revealed a dramatic reduction of radial glia and fiber density compared to control (G). Remaining GFAP+ cells in ΔStumpy mice had abnormal morphology characterized by disorganized and misoriented processes. (I and J) DAPI-nuclear staining showed decreased cell density in both the SGZ and GCL of mutants (J) compared to control (I).
ΔStumpy mice exhibit hydrocephalus (14), and we initially thought that this could be in part responsible for the abnormalities observed. However, we recovered several “control” idiopathic spontaneously hydrocephalic mice (NestinCre−/Stumpy+/+) with a comparable severity of hydrocephalus, but which lacked any significant malformations in the cerebellum or hippocampus (Fig. S3). In addition, we encountered several other spontaneously hydrocephalic mice from different strains with comparable levels of hydrocephalus (Fig. S3). None of these animals exhibited the characteristic anatomical phenotypes observed in ΔStumpy mice. Therefore, loss of Stumpy has specific consequences on the number and morphology of neuronal precursors in the hippocampus and cerebellum that was independent of hydrocephalus.
In situ hybridization (ISH) for Stumpy in the postnatal hippocampus showed a fairly ubiquitous expression pattern (Fig. S4 A and B). Using an antibody against Stumpy (14), we observed Stumpy colocalization with the basal body (Fig. S4 C and D). Using the marker adenylyl cyclase III (ACIII), which localizes to the primary cilium (5), cilia were observed on many cells in the control DG and were completely absent in ΔStumpy mice (Fig. S4 E and F′). Somatostatin Receptor 3, another marker of neuronal cilia, yielded similar results (Fig. S5). Also, ultrastructural examination of radial glia confirmed the lack of ciliary axonemes in ΔStumpy mice (Fig. S6). Therefore, loss of stumpy leads to an absence of ciliary axonemes in the hippocampus.
Astroglial cells, the major neural precursors in hippocampus, were markedly disrupted in ΔStumpy (Fig. 1 E–H). To determine whether cilia were present on these precursors, we used an inducible GFP-labeling method to mosaically label them in a Golgi-like manner (Fig. 2A). We identified ACIII+ cilia on these neurogenic, astrocyte-like neural precursors (ALNPs) in the hilus and subgranular zone (SGZ) (Fig. 2 B and B″). Interestingly, these cilia were morphologically distinct—i.e., significantly shorter—from neuronal cilia (2.6 ± 0.5 μm in glial cells vs. 8.2 ± 1.7 μm in neurons; P < 0.05 [Student's t test]). These cilia were present on both the radial and nonradial ALNP populations and were often recessed into the cell membrane (Fig. 2 C–F). Neuronal cilia normally protruded with no evidence of membrane invagination (data not shown). We confirmed the findings of glial cilia in vitro using two different radial glia-like cell lines: C6-R cells (derived from rat glioma cells) and bipolar adult neural progenitor cells harvested using published techniques (Fig. S7) (15). Both cell lines use Shh signaling to maintain the stem cell population (16, 17). These results suggest that cilia on ALNPs could play an important role in regulating their subsequent proliferation and/or differentiation into neurons.
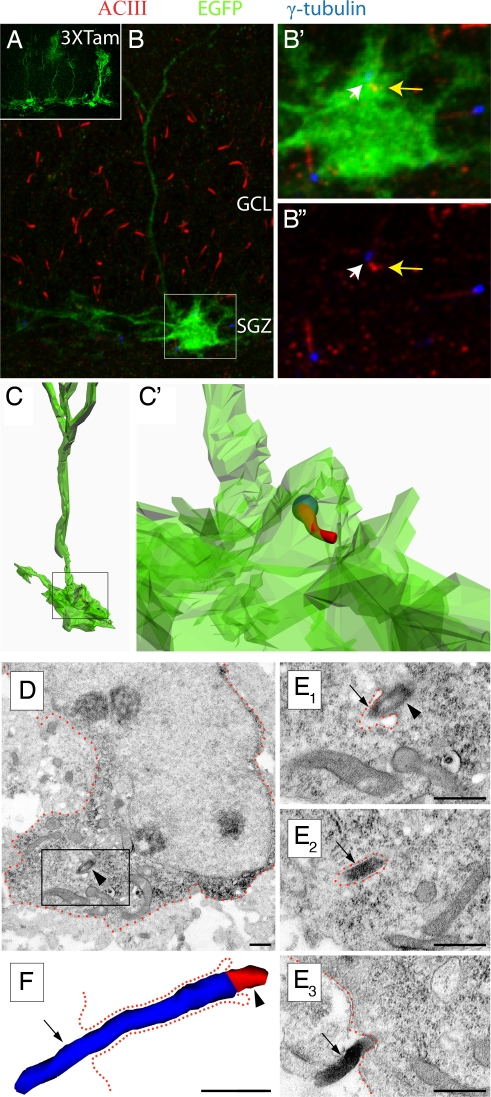
Fig. 2.
Cilia are present on hippocampal ALNPs. (A) GFAP-CRE-EGFP transgenic mice were injected with tamoxifen for three days and sacrifed one day after the last injection. An example of mosaic labeling of glia with EGFP is shown. (B) EGFP-expressing radial glial cell (boxed) in the subgranular zone (SGZ) in conjunction with immunstaining for ACIII (red) and γ-tubulin (blue). Cilia were located in both GCL and SGZ. (B′) Higher magnification of the inset in B. A cilium was observed extending from the EGFP+ cell expressing both γ-tubulin (white arrow) and ACIII (yellow arrow). (B″) The inset in B showing γ-tubulin (white arrow) and ACIII (yellow arrow). Note that the cilia length is much shorter than overlying GCL cells. (C, C′) Z-stack data were used to reconstruct the cell in B and the location of the cilum within the cell membrane (C′). The spatial resolution of this technique is limited by the technology. Nevertheless, the cilium appears to be largely enveloped in the EGFP+ membrane. (D) An astrocyte glial cell harboring a cilium in the SGZ of a P7 wild-type mouse. Low power electron micrograph of a GFAP+ cell detected with diffuse anti-GFAP DAB immunoprecipitation in the cytoplasm. The framed area is depicted in serial high power micrographs in E. (E) Examples of the basal body (arrowhead) and the ACIII+ axoneme (arrow) detected with black Ni-intensified DAB-immunoprecipitation. (F) Three-dimensional-reconstruction of the cilium showing the basal body (red; arrowhead) and the axoneme (blue; arrow). The cilium was followed in 12 consecutive serial sections until its termination. The total length of the axoneme (∼1.7 μm) was measured in 3D reconstruction. The cell membrane is approximately outlined by green dotted lines. Scale bars, 0.5 μm.
Stumpy Mutants Have Defects in Neuronal Precursor Proliferation and Neurogenesis Associated with Perturbed Shh Signaling.
Analysis of P0 hippocampi indicated that proliferation of DG precursors was already altered in ΔStumpy mutants, as the DG appeared smaller and had fewer dividing precursor cells (Fig. S8 A and B). When we examined the proliferation of Sox2+ glia later at P13, ΔStumpy mice showed fewer Sox2+ cells and a more random distribution when compared with controls (Fig. 1 A, B, E, and F). There was less proliferation overall as determined by the cell cycle marker Ki67 expression in ΔStumpy mice (Fig. S9 A and B). Notably, fewer proliferating Sox2+ cells were present, as determined by colocalization with Ki67 (∼52 ± 5% of Ki67+ cells were Sox2+ in controls vs. ∼70 ± 8% Sox2+/Ki67+ cells in ΔStumpy mice; P < 0.05 [Student's t test]) (Fig. S9 A and B). However, it should be noted that there appeared to be fewer weakly Sox2+ cells that colocalized with Ki67 (Fig. S9 A and B)—perhaps indicative of a decrease in proliferating transit amplifying cells (18). Neurogenesis, in the form of transit-amplifying cells and migrating neurons, was similarly altered in Stumpy mutants. In particular, we noted ectopic Ascl1, and Tbr2 expression—early markers of neurogenesis (19) (Fig. S9 C and D″). Immature neurons, stained with doublecortin (Dcx), also displayed abnormal morphology (Fig. S9 C′ and D′). Taken together, Stumpy mutants exhibit profound alterations in early (P0) and later proliferation (P13), leading to subsequent deficiencies in neurogenesis.
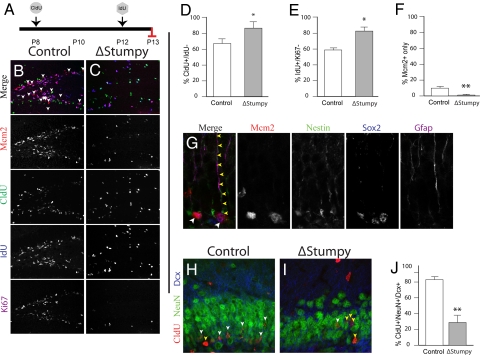
To assay where the primary defect in neurogenesis lay, we birthdated two separate populations of S-phase cells with the DNA synthesis markers chlorodeoxyuridine (CldU) and iododeoxyuridine (IdU). The CldU was administered at P8, and IdU was given at P12 and animals were killed at P13 (Fig. 3A). In controls, hippocampal proliferation was largely confined to the SGZ. However, in ΔStumpy mutants, proliferation was diminished in the SGZ and ectopic proliferating cells were more abundant (Fig. 3 B and C). To determine whether this effect was because of abnormal cell cycle exit, we examined the proportion of cells labeled with CldU but not IdU and noted an increase in ΔStumpy mice, indicating an increase in postmitotic cells at this time (Fig. 3D). When looking at cell cycle exit after 24 h (i.e., the percentage of Idu+ cells that were Ki67−), we found a similar increase in ΔStumpy mutants (Fig. 3E). Importantly, when using the marker Mcm2 for proliferating and quiescent precursor cells (20), we could use combinatorial staining for this protein with Ki67, IdU, and CldU to highlight the slowly dividing, quiescent population (i.e., cells that are Mcm2+ but Ki67−, IdU−, and CldU−). We noted a drastic depletion of this quiescent population in ΔStumpy mice (Fig. 3F)—consistent with a lack of self-renewal of this population. Mcm2+ cells in controls frequently colocalized with Nestin, Sox2, and Gfap, and often exhibited a stellate or radial morphology (Fig. 3G). Lastly, neurogenesis, as determined by colocalization of NeuN, CldU, and Dcx (Fig. 3 H and I), was notably diminished in ΔStumpy mice (Fig. 3J). Thus, hippocampal defects in ΔStumpy mice appear to be because of a smaller quiescent precursor population and increased cell cycle exit, leading to a net reduction in DG neurons.
Fig. 3.
Altered cell cycle dynamics, proliferation and differentiation. (A) Control or ΔStumpy mice were injected with CldU at P8, IdU on P12 and killed at P13. (B and C) Immunostaining for Mcm2 (red), Ki67 (magenta), Idu (blue), and Cldu (green) in control showed predominant labeling within the SGZ and the GCL. Conversely, ΔStumpy mice exhibited diffusely distributed proliferating cells and an overall reduction of proliferating cells. Arrows in B point to the Mcm2+ population that does not colocalize with the other proliferative markers, indicative of slowly dividing cells in G1 of the cell cycle. Note the complete lack of this population in C. (D) The percentage of cells that were CldU+ and Idu− was higher in ΔStumpy brains, indicating that less of the proliferating pool at P8 is in the cell cycle at P12. (E) The percentage of cells that were Idu+ and Ki67− at P13. In ΔStumpy mice, the proportion of cells that exited the cell cycle after 24 h is increased compared with controls. (F) The percentage of cells that were Mcm2+ and negative for Ki67, CldU, and IdU at P13. This slowly dividing stem/precursor pool is largely depleted in ΔStumpy mice. (G) Nestin+/Gfap+/Sox2+ cells—radial and nonradial glia—make up a large population of the Mcm2 population in controls (white arrowheads). Note the attached Nestin+/Gfap+ radial process (yellow arrowheads). (H–J) The reduction of dividing cells [labeled at P8 with CldU (red)] results in a reduced number of DCX (blue) and NeuN- (green) positive neurons (white arrowheads) in ΔStumpy mutant DG (I) compared to control (H)—quantified in (J). Non-neuronal CldU+ cells are pointed out by yellow arrowheads. *P < 0.05; **P < 0.001 (Student's t test).
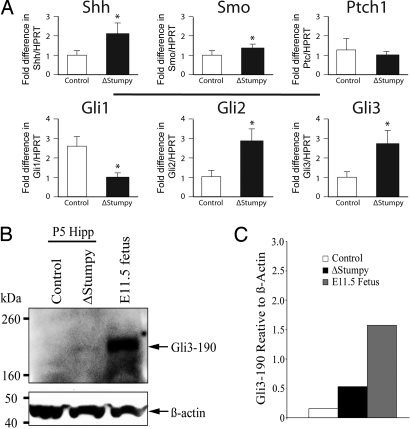
The similar DG phenotypes between ΔStumpy mice and NestinCre; Smofl/fl mice (10), and the observed proliferation defects seemed consistent with a defect in the Shh pathway. Thus, we next examined expression of critical Shh signaling mediators. We screened hippocampal mRNA for Shh, Smo, Ptch, and Gli1–3 (relative to the levels of Hprt1) between control and ΔStumpy mice by qRT-PCR. We found a significant decrease in Gli1 mRNA levels (Fig. 4A), an indicator of reduced Shh signaling (13). Interestingly, Shh levels were increased in ΔStumpy hippocampus, suggesting a compensatory mechanism or potential alterations in the processing of Gli3 (1, 4, 21–23). We also found that Smo, Gli2, and Gli3 were up-regulated compared to control (Fig. 4A).
Fig. 4.
Altered mRNA levels of Shh pathway genes and changes in Gli3 processing. (A) Expression levels of Shh pathway genes in hippocampus using qRT-PCR. Levels of Shh, Smo, Ptc, and Gli1–3 were normalized to levels of HPRT. *P < 0.05 (Student's t test). (B) Western blot showing the levels of full-length Gli3 (∼190 kDa) in P5 hippocampus (Hipp) of control and ΔStumpy compared to E11.5 fetus. Note the absence of a detectable band in the control lane. Shown below is the β-actin loading control. (C) The blot in (B) was repeated in triplicate and the graph shows the average intensity of Gli3-190 signal relative to β-actin signal for the indicated groups.
Gli2 and Gli3 regulate the subsequent expression of Gli1 (21, 24). However, both were increased in ΔStumpy hippocampus; yet, Gli1 mRNA was decreased. Thus, the expression of Gli2 and Gli3 occur in a cilia-independent manner in contrast to Gli1. Strikingly, Shh mRNA was also increased in the ΔStumpy hippocampus. Gli3, in particular, can be processed from a full-length activator form (∼190 kDa) into a shorter, repressor protein (∼83 kDa), which represses transcription of Gli1 and Shh (22). This processing is thought to be cilia-dependent and allows for the proper cellular response to the presence or absence of Shh. Thus, we hypothesized that improper Gli3 processing could underlie aspects of the ΔStumpy phenotype. Gli3 in the postnatal hippocampus was barely detectable compared to robust E11.5 expression (Fig. 4 B and C). Nevertheless, we noted a prominent increase in Gli3 activator in ΔStumpy mice compared with control (Fig. 4 B and C), leading to an altered ratio of activator to repressor forms (data not shown). These results are consistent with previous reports that cilia are necessary for the proper cleavage of Gli3 (4, 25). Our results show conspicuous alterations in Shh signaling and Gli processing in mice lacking Stumpy.
Expression of Shh Signaling-Associated Molecules at the Primary Cilium Regulates Proliferation.
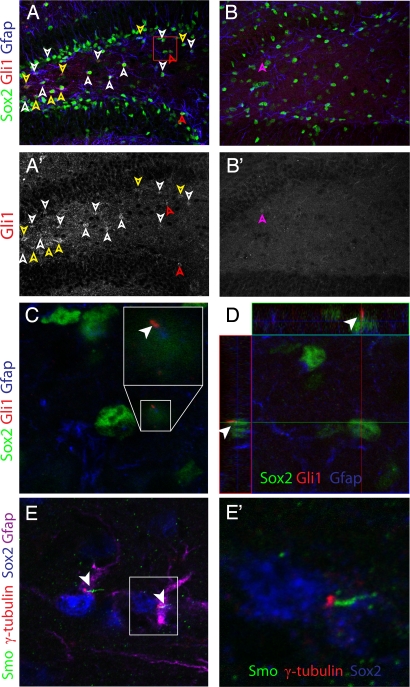
Ptch1, Smo, and Gli1 are expressed in a pattern consistent with a role in neurogenesis in the SGZ (Fig. S10), in line with previous reports (9, 10, 13). Furthermore, a recent paper detailing the differential transcriptome of specific cell types in the brain indicated that Shh signaling is largely confined to astrocytes (26). Indeed, Gli1 immunostaining was largely localized to glial nuclei, and, to a lesser extent, glial cytoplasm in the SGZ and hilus (Fig. 5A and A′). Notably, and consistent with our qRT-PCR results, Gli1 was barely detectable in ΔStumpy mice (Fig. 5 B and B′). On closer inspection, rare cells in the control SGZ and hilus exhibited Gli1 enrichment in cilia-like protrusions from the cell body (Fig. 5 C and D), supporting previous observations of Gli1 within primary cilia (25). Furthermore, cells which had notable enrichment of ciliary Gli1 and low but detectable levels of nuclear Gli1 were observed as well (data not shown), possibly reflecting an active, dynamic change in subcellular localization of Gli1 which would be expected following Smo de-repression. Using an antibody specific for Smo (27), we found that Smo also localized to rare, scattered, short, glial cilia in the SGZ and hilus (Fig. 5E–E′). Neither Gli1 nor Smo were found to localize to neuronal cilia (data not shown).
Fig. 5.
Localization of Shh molecules to ALNP cilia. (A and B) Immunostaining for Gli1, Sox2 and GFAP in control (A) and ΔStumpy (B) mutants. Gli1 largely colocalized with both Sox2 and GFAP in control. Gli1 immunostaining was reduced in ΔStumpy brains. White arrowheads denote Gli1 cytoplasm/nuclei. Yellow arrowheads denote cytoplasmic Gli1. Red arrowheads denote presumptive ciliary Gli1. Purple arrowhead in B and B′ denotes a single, faint Gli1+ nucleus in a Sox2− cell. Gli1 immunostaining channel from A and B is shown from control (A′) and ΔStumpy (B′) tissue. (C and D) Higher magnification of boxed region in (A). Shown in (C) is a single optical section from the orthographic view shown in (D). The Gli1 enrichment (white arrowhead) resembles localization to a cilium. (E, E′) Immunostaining of Smo (green), γ-tubulin (red), Sox2 (blue), and Gfap (magenta) in a pair of hilar astrocytes near the SGZ. Smo appears enriched in short cilia charateristic of ALNPs. (E′) is a higher magnification of boxed region in E.
The localization of Shh-associated molecules to cilia in the neurogenic regions of hippocampus together with the reduced proliferation of ALNPs, and lack of Gli1 immunostaining in ΔStumpy hippocampus suggests that Shh may act through cilia to regulate proliferation. We tested this in P8 hippocampal slice cultures and asked whether exogenously applied Shh could alter proliferation in the DG. Shh-treated cultures showed a marked hilar and SGZ proliferative phenotype (data not shown)—consistent with previous reports in this and other brain regions (9, 10, 17, 28). In contrast, Shh application to ΔStumpy hippocampi failed to induce proliferation when compared with ΔStumpy controls (data not shown). Thus, Shh appears to act through cilia on ALNPs to control the proliferation of neuronal precursors.
Discussion
Here, we show that cilia are critical regulators of Shh signaling in postnatal precursor cells and therefore participate in orchestrating postnatal forebrain development and stem/precursor cell maintenance. The absence of cilia and subsequent alteration of Shh signaling leads to defective hippocampal morphogenesis and dysregulation of mitotic activity. Our interpretation is that stumpy, by way of its requirement for ciliogenesis, enables Shh signaling at the primary cilium and subsequent neuronal precursor proliferation and postnatal neurogenesis. Whether stumpy directly participates in the Shh signaling pathway, or in the cell cycle; or whether there is some cilia-independent Shh-signaling in this region is unknown.
Radial glia are considered to be the primary neuronal precursors during embryonic CNS development, and these cells typically project a primary cilium into the ventricle during interphase (29). Whether secreted Shh acts on these cells remains unclear, as most brain regions are reported to form normally in NestinCre; Smofl/fl mutants (10). However, postnatal radial glia transition into ALNPs, protoplasmic astrocytes, and ependymal cells (in addition to several other neural cell types) (29). Importantly, these transitions for SGZ and SEZ ALNPs coincide with their responsiveness to Shh (10, 12, 13). This is supported by SGZ and SEZ abnormalities following decreased precursor expansion and subsequent progenitor depletion primarily in the first two postnatal weeks in NestinCre; Smofl/fl mutants (10), and the activity of Shh- and Smo-responsive progenitors in replenishing of the neurogenic niche after anti-mitotic treatments (12, 13). The increased hippocampal Shh we observed may reflect a compensatory response to the loss of cilia (Fig. 4A). However, we also found altered Gli3 processing in ΔStumpy mice (Fig. 4B), which may underlie these changes in Shh levels. Here, we link the proper processing of Gli3 in the cilium to neural precursor/stem cell proliferation and self-renewal.
Our data demonstrate that, in the absence of cilia, there is a dramatic diminution in Shh signaling, decreased early proliferation at P0, and a consequent loss of quiescent precursor cells later at P13. The smaller size of the DG at P0, the near complete loss of nuclear Gli1 in the SGZ/hilus, and the loss of slowly dividing Mcm2+ cells in ΔStumpy at P13 support this. An understanding of cilia-mediated Shh signaling and its consequences for cell proliferation have profound clinical implications beyond hippocampal neurogenesis. The CNS cancer stem cell is Shh-responsive (16), and Shh-related mutations have been linked to meduloblastoma (30). Thus, a better understanding of cilia function in neural precursor cells may not only serve to illuminate the physiologic mechanisms of precursor proliferation and postnatal neurogenesis, but may also yield novel insights into CNS oncogenesis.
Note.
After completing these studies, Han et al. (31) reported that Shh signaling is required for DG proliferation via cilia. Supporting and complementing our findings, their work shows that mice lacking other cilia genes (e.g., Kif3a and Ift88) have similar defects in ALNP morphology and numbers. Further, an attempt to rescue DG proliferation by restoring Shh signaling (by constitutive activation of Smo) failed to restore neurogenesis in cilia mutants, indicating the necessity of an intact, functional cilium. Additionally, Spassky et al. (32) showed that application of Shh caused massive induction of EGL precursor proliferation that was absent in Kif3a mutant mice—as we observed in the DG—strengthening the link between Shh signaling, primary cilia, and postnatal neurogenesis.
Materials and Methods
Mice and Genotyping.
Stumpy mutant mice were generated as previously described (14). Briefly, a floxed stumpy allele was deleted in the presence of Cre under the control of the Nestin promoter. Controls consisted of Nestin-Cre; Stumpyfl/+ mice unless otherwise noted. GCE; CAG-CAT-GFP mice for mosaic labeling were generated as described previously (19). Genotyping for all strains was carried out as described previously (14, 19). All animal protocols were in accordance with Yale University and IACUC guidelines.
Immmunostaining.
Postnatal mice were perfused intracardially with 1X PBS followed by 4% PFA. Brains were dissected and fixed overnight in 4% PFA, rinsed, cryoprotected and frozen over liquid N2. Twenty μm cryosections were sliced on a cryostat. Standard immunostaining procedures were used for most antibodies and appropriate secondary-conjugated antibodies. For BrdU/CldU/IdU immunostaining, sections were pretreated in 2N HCL for 15 min.
Antibodies.
The following antibodies were used: mouse anti-acetylated alpha tubulin (1:1000; Sigma), mouse anti-gamma tubulin (1:1000; Sigma), rabbit anti-gamma tubulin (1:1000; Sigma), rabbit anti-adenylyl cyclase III (1:500; Santa Cruz Biotechnology), goat anti-Mcm2 (1:400; Santa Cruz), mouse anti-GFAP (1:1000; Sigma), rat anti-BrdU (for CldU detection; 1:250; Accurate), mouse anti-brdU (also for IdU detection; 1:250; Becton Dickinson), chicken anti-EGFP (1:5000; Abcam), rabbit anti-Ki67 (1:250; Vector), mouse anti-Calbindin (1:1000; Swant), goat anti-DCX (1:500; Santa Cruz Biotechnology), anti-Gli1 (1:100; Novus Biologicals), mouse anti-MASH1 (aka Ascl1, BD PharMingen), mouse anti-NeuN (1:1000; Chemicon), rabbit anti-S100β (1:1000; Sigma), goat anti-SOX2 (1:500; Santa Cruz Biotechnology), rabbit anti-SSR3 (1:2000; Gramsch), rabbit anti-Smo (1:200; Lifespan Biosciences), rabbit anti-Tbr2 (1:20000; Chemicon). Rabbit anti-Stumpy polyclonal antibody was generated as previously described (1:2000) (14).
Details of all additional methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank M. Pappy for excellent experimental assistance; M. Li and F.M. Vaccarino for mice; and K. Hashimoto-Torii, M. Torii, R. Rasin, and members of the Rakic and Sestan Labs for helpful discussions, technical support, and comments. We thank A. Alvarez-Buylla for discussions and for sharing findings with us before submission. This study was supported by the U.S. National Institutes of Health and the Kavli Institute. T.T. is supported by an NIH/NIA “Pathway to Independence” Award (1K99AG029726–01 and 4R00AG029726–02) and an Alzheimer's Association grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804558105/DCSupplemental.
References
- 1.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawamoto K, et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–632. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- 3.Davenport JR, et al. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berbari NF, Bishop GA, Askwith CC, Lewis JS, Mykytyn K. Hippocampal neurons possess primary cilia in culture. J Neurosci Res. 2007;85:1095–1100. doi: 10.1002/jnr.21209. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs JL, Schwark HD. Neuronal primary cilia: A review. Cell Biol Int. 2004;28:111–118. doi: 10.1016/j.cellbi.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 8.Huangfu D, et al. Hedgehog signaling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 9.Palma V, et al. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machold R, et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron. 2003;39:937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- 11.Rakic P. Limits of neurogenesis in primates. Science. 1985;227:1054–1056. doi: 10.1126/science.3975601. [DOI] [PubMed] [Google Scholar]
- 12.Balordi F, Fishell G. Mosaic Removal of Hedgehog Signaling in the Adult SVZ Reveals That the Residual Wild-Type Stem Cells Have a Limited Capacity for Self-Renewal. J Neurosci. 2007;27:14248–14259. doi: 10.1523/JNEUROSCI.4531-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- 14.Town T, et al. The stumpy gene is required for mammalian ciliogenesis. Proc Natl Acad Sci USA. 2008;105:2853–2858. doi: 10.1073/pnas.0712385105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray J, Gage FH. Differential properties of adult rat and mouse brain-derived neural stem/progenitor cells. Mol Cell Neurosci. 2006;31:560–573. doi: 10.1016/j.mcn.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Bar EE, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai K, Kaspar BK, Gage FH, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- 18.Komitova M, Eriksson PS. Sox-2 is expressed by neural progenitors and astroglia in the adult rat brain. Neurosci Lett. 2004;369:24–27. doi: 10.1016/j.neulet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 19.Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci USA. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maslov AY, Barone TA, Plunkett RJ, Pruitt SC. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. J Neurosci. 2004;24:1726–1733. doi: 10.1523/JNEUROSCI.4608-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: An information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 23.Buscher D, Bosse B, Heymer J, Ruther U. Evidence for genetic control of Sonic hedgehog by Gli3 in mouse limb development. Mech Dev. 1997;62:175–182. doi: 10.1016/s0925-4773(97)00656-4. [DOI] [PubMed] [Google Scholar]
- 24.Lei Q, et al. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development. 2004;131:3593–3604. doi: 10.1242/dev.01230. [DOI] [PubMed] [Google Scholar]
- 25.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbit C, et al. Vertebrate smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 28.Mullor JL, Dahmane N, Sun T, Ruiz I, Altaba A. Wnt signals are targets and mediators of Gli function. Curr Biol. 2001;11:769–773. doi: 10.1016/s0960-9822(01)00229-9. [DOI] [PubMed] [Google Scholar]
- 29.Spassky N, et al. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 31.Han YG, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 32.Spassky N, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.