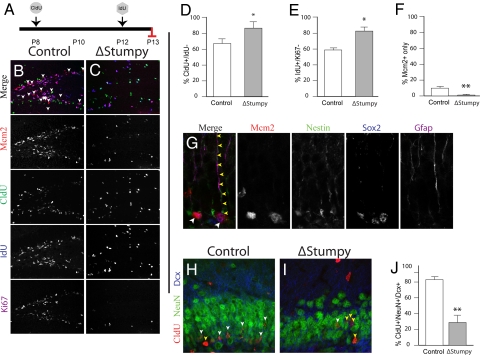
Fig. 3.
Altered cell cycle dynamics, proliferation and differentiation. (A) Control or ΔStumpy mice were injected with CldU at P8, IdU on P12 and killed at P13. (B and C) Immunostaining for Mcm2 (red), Ki67 (magenta), Idu (blue), and Cldu (green) in control showed predominant labeling within the SGZ and the GCL. Conversely, ΔStumpy mice exhibited diffusely distributed proliferating cells and an overall reduction of proliferating cells. Arrows in B point to the Mcm2+ population that does not colocalize with the other proliferative markers, indicative of slowly dividing cells in G1 of the cell cycle. Note the complete lack of this population in C. (D) The percentage of cells that were CldU+ and Idu− was higher in ΔStumpy brains, indicating that less of the proliferating pool at P8 is in the cell cycle at P12. (E) The percentage of cells that were Idu+ and Ki67− at P13. In ΔStumpy mice, the proportion of cells that exited the cell cycle after 24 h is increased compared with controls. (F) The percentage of cells that were Mcm2+ and negative for Ki67, CldU, and IdU at P13. This slowly dividing stem/precursor pool is largely depleted in ΔStumpy mice. (G) Nestin+/Gfap+/Sox2+ cells—radial and nonradial glia—make up a large population of the Mcm2 population in controls (white arrowheads). Note the attached Nestin+/Gfap+ radial process (yellow arrowheads). (H–J) The reduction of dividing cells [labeled at P8 with CldU (red)] results in a reduced number of DCX (blue) and NeuN- (green) positive neurons (white arrowheads) in ΔStumpy mutant DG (I) compared to control (H)—quantified in (J). Non-neuronal CldU+ cells are pointed out by yellow arrowheads. *P < 0.05; **P < 0.001 (Student's t test).