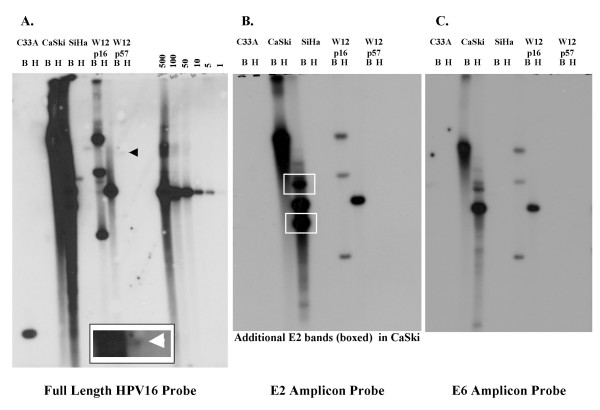
Figure 3.
Southern analysis of HPV16 copy number and physical state in the cervical keratinocyte cell lines used to validate the NA6 qPCR method. The three panels represent blots probed with full length HPV16 (A), the HPV16 E2 amplicon (B) and the HPV16 E6 amplicon (C). The lanes marked B are BamH1 digests, while the lanes marked H are Hind III digests. The right hand lanes in Panel A show copy number controls for estimates of viral load, ranging from 500 to 1 copies of full length HPV16 per 5 μg of normal peripheral blood lymphocyte gDNA. C33A is HPV16 negative, CaSki contains integrated HPV16 at high copy, SiHa contains integrated HPV16 at low copy, W12.Ser1p16 contains episomal HPV16, and W12.Ser1p57 contains a single HPV16 integrant (arrowed). The inset at the bottom of Panel A shows the faint signal produced by the HPV16 integrant in W12.Ser1p57 following prolonged (72 hr) exposure. For CaSki cells, the signal produced with the E2 probe (Panel B) is considerably greater than that with the E6 probe (Panel C) and additional E2 bands are present (boxed in Panel B), indicative of multiple integration sites.