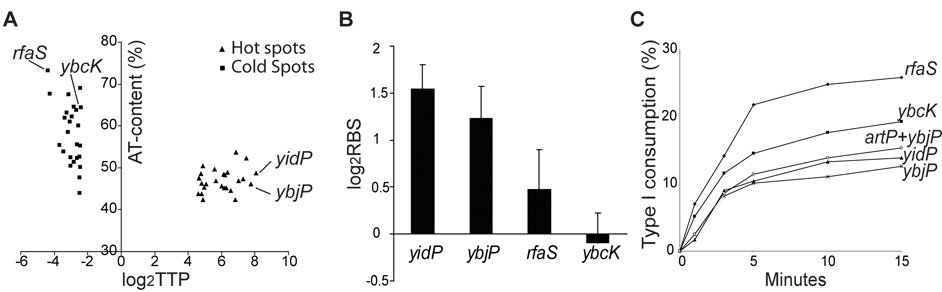
Figure 2.
A. A/T content of 25 hot and cold Mu target genes in E. coli. The log2TTP data is taken from Table 1 in Ref. 8, and A/T content of each gene is based on the E. coli K12 genome sequence (NC_000913). Indicated genes were used for further study. B. The relative binding strength (RBS) of MuB on hot and cold genes in vivo. MuB binding was measured by ChIP assays using MuB antibody (see Methods). Bound DNA was amplified by PCR and quantitated. RBS or relative binding strength is the ratio of specific to non-specific binding. The binding data are an average of three repeats performed for the following regions showing the strongest binding within each gene: yidP (49% A/T), 351–717bp; ybjP (46% A/T), 210–516 bp; rfaS (73% A/T), 601–936 bp; ybcK (64% A/T), 351–700bp. Student-t tests were performed, and the p-values (p<0.05) suggest that MuB binding to hot targets genes was significantly stronger than that to cold target genes. C. In vitro target efficiency of hot and cold in vivo targets. Strand transfer was initiated by adding equal amounts of the cleaved Mu complex assembled on pSP104 to 2-fold molar excess of PCR-amplified linear target DNA derived from the indicated genes. Reaction aliquots taken at various times were run on an agarose gel, and donor or target consumption measured by quantifying the intensity of the appropriate DNA bands. Target sizes are as follows: yidP (717 bp), ybjP (516 bp), rfaS (936 bp), ybcK (1527 bp), artP + ybjP (1452 bp).