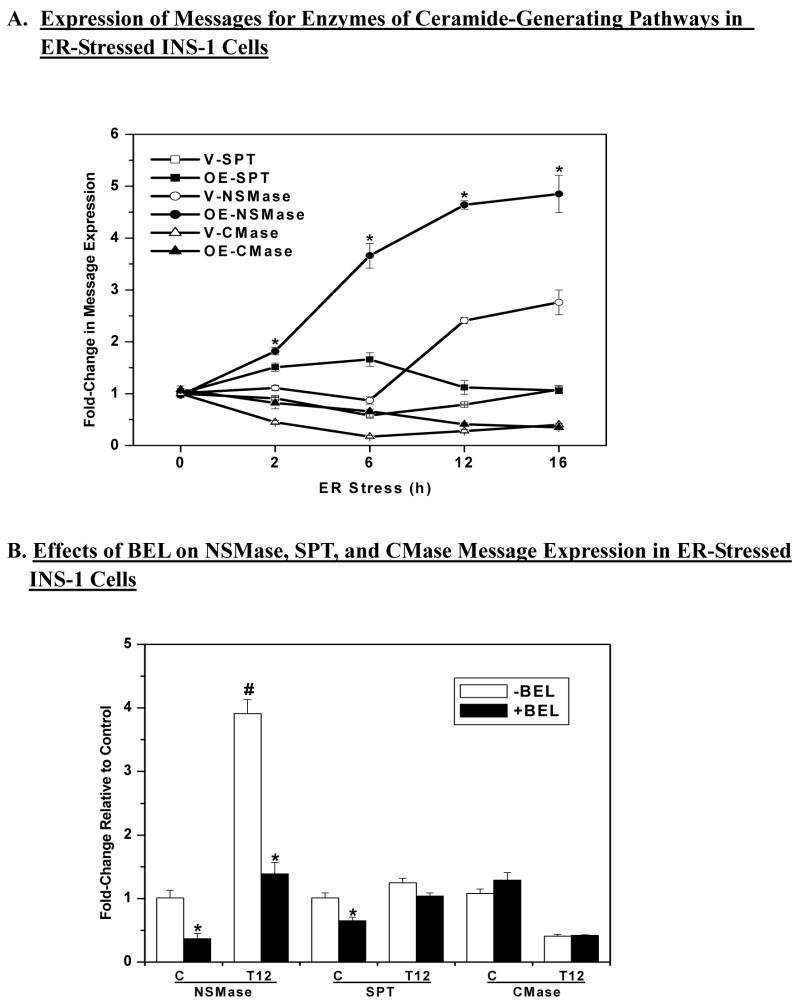
Figure 3. ER stress induced expression of neutral sphingomyelinase (NSMase) message in INS-1 cells ± BEL.
INS-1 cells were treated with either vehicle (DMSO, C) or with thapsigargin (T, 1 μM) and cultured for up to 16 h in the absence or presence of BEL (1 μM). Total RNA was prepared from the cells at various times for quantitative (Q) RT-PCR analyses for enzymes that participate in the generation of ceramides via the de novo pathway (SPT, serine palmitoyl transferase), sphingomyelin hydrolysis (NSMase), and inhibition of ceramide degradation (CMase, ceramidase). A. QRT-PCR analyses of SPT, NSMase, and CMase (n = 4). (*OE-NSMase group significantly different from V-NSMase group, p < 0.05). B. QRT-PCR analyses of NSMase, SPT, and ceramidase ± BEL in control cells and 12 h after induction of ER stress (T12) (n=4). (*BEL-treated group significantly different from corresponding untreated group, p < 0.05. #Untreated NSMase group at 12 h significantly different from other NSMase groups, p < 0.05.).